Host gene expression classifiers diagnose acute respiratory illness etiology
- PMID: 26791949
- PMCID: PMC4905578
- DOI: 10.1126/scitranslmed.aad6873
Host gene expression classifiers diagnose acute respiratory illness etiology
Abstract
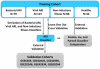
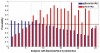
Acute respiratory infections caused by bacterial or viral pathogens are among the most common reasons for seeking medical care. Despite improvements in pathogen-based diagnostics, most patients receive inappropriate antibiotics. Host response biomarkers offer an alternative diagnostic approach to direct antimicrobial use. This observational cohort study determined whether host gene expression patterns discriminate noninfectious from infectious illness and bacterial from viral causes of acute respiratory infection in the acute care setting. Peripheral whole blood gene expression from 273 subjects with community-onset acute respiratory infection (ARI) or noninfectious illness, as well as 44 healthy controls, was measured using microarrays. Sparse logistic regression was used to develop classifiers for bacterial ARI (71 probes), viral ARI (33 probes), or a noninfectious cause of illness (26 probes). Overall accuracy was 87% (238 of 273 concordant with clinical adjudication), which was more accurate than procalcitonin (78%, P < 0.03) and three published classifiers of bacterial versus viral infection (78 to 83%). The classifiers developed here externally validated in five publicly available data sets (AUC, 0.90 to 0.99). A sixth publicly available data set included 25 patients with co-identification of bacterial and viral pathogens. Applying the ARI classifiers defined four distinct groups: a host response to bacterial ARI, viral ARI, coinfection, and neither a bacterial nor a viral response. These findings create an opportunity to develop and use host gene expression classifiers as diagnostic platforms to combat inappropriate antibiotic use and emerging antibiotic resistance.
Copyright © 2016, American Association for the Advancement of Science.
Figures


References
-
- WHO [accessed on Nov 8, 2011];Mortality and burden of disease: Cause-specific mortality, 2008: WHO regions. Global Health Observatory Data Repository. 2011 Available at: http://apps.who.int/ghodata/
-
- Shapiro DJ, Hicks LA, Pavia AT, Hersh AL. Antibiotic prescribing for adults in ambulatory care in the USA, 2007-09. The Journal of antimicrobial chemotherapy. 2014;69:234–240. - PubMed
-
- Gould IM. Antibiotic resistance: the perfect storm. International Journal of Antimicrobial Agents. 2009;34(Supplement 3):S2–S5. - PubMed
-
- Kim JH, Gallis HA. Observations on spiraling empiricism: its causes, allure, and perils, with particular reference to antibiotic therapy. Am J Med. 1989;87:201–206. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

