The ubiquitin ligase Ubr4 controls stability of podocin/MEC-2 supercomplexes
- PMID: 26792178
- PMCID: PMC4787903
- DOI: 10.1093/hmg/ddw016
The ubiquitin ligase Ubr4 controls stability of podocin/MEC-2 supercomplexes
Abstract
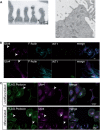
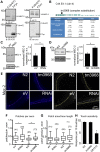
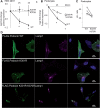
The PHB-domain protein podocin maintains the renal filtration barrier and its mutation is an important cause of hereditary nephrotic syndrome. Podocin and its Caenorhabditis elegans orthologue MEC-2 have emerged as key components of mechanosensitive membrane protein signalling complexes. Whereas podocin resides at a specialized cell junction at the podocyte slit diaphragm, MEC-2 is found in neurons required for touch sensitivity. Here, we show that the ubiquitin ligase Ubr4 is a key component of the podocin interactome purified both from cultured podocytes and native glomeruli. It colocalizes with podocin and regulates its stability. In C. elegans, this process is conserved. Here, Ubr4 is responsible for the degradation of mislocalized MEC-2 multimers. Ubiquitylomic analysis of mouse glomeruli revealed that podocin is ubiquitylated at two lysine residues. These sites were Ubr4-dependent and were conserved across species. Molecular dynamics simulations revealed that ubiquitylation of one site, K301, do not only target podocin/MEC-2 for proteasomal degradation, but may also affect stability and disassembly of the multimeric complex. We suggest that Ubr4 is a key regulator of podocyte foot process proteostasis.
© The Author 2016. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

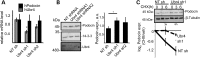


References
-
- Boute N., Gribouval O., Roselli S., Benessy F., Lee H., Fuchshuber A., Dahan K., Gubler M.C., Niaudet P., Antignac C. (2000) NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat. Genet., 24, 349–354. - PubMed
-
- Tory K., Menyhárd D.K., Woerner S., Nevo F., Gribouval O., Kerti A., Stráner P., Arrondel C., Huynh Cong E., Tulassay T. et al. (2014) Mutation-dependent recessive inheritance of NPHS2-associated steroid-resistant nephrotic syndrome. Nat. Genet., 46, 299–304. - PubMed
-
- Pavenstädt H., Kriz W., Kretzler M. (2003) Cell biology of the glomerular podocyte. Physiol. Rev., 83, 253–307. - PubMed
-
- Huber T.B., Schermer B., Benzing T. (2007) Podocin organizes ion channel-lipid supercomplexes: implications for mechanosensation at the slit diaphragm. Nephron Exp. Nephrol., 106, e27–e31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

