The P1N-PISPO trans-Frame Gene of Sweet Potato Feathery Mottle Potyvirus Is Produced during Virus Infection and Functions as an RNA Silencing Suppressor
- PMID: 26792740
- PMCID: PMC4794657
- DOI: 10.1128/JVI.02360-15
The P1N-PISPO trans-Frame Gene of Sweet Potato Feathery Mottle Potyvirus Is Produced during Virus Infection and Functions as an RNA Silencing Suppressor
Abstract
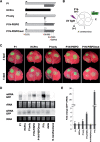
The positive-sense RNA genome of Sweet potato feathery mottle virus (SPFMV) (genus Potyvirus, family Potyviridae) contains a large open reading frame (ORF) of 3,494 codons translatable as a polyprotein and two embedded shorter ORFs in the -1 frame: PISPO, of 230 codons, and PIPO, of 66 codons, located in the P1 and P3 regions, respectively. PISPO is specific to some sweet potato-infecting potyviruses, while PIPO is present in all potyvirids. In SPFMV these two extra ORFs are preceded by conserved G2A6 motifs. We have shown recently that a polymerase slippage mechanism at these sites could produce transcripts bringing these ORFs in frame with the upstream polyprotein, thus leading to P1N-PISPO and P3N-PIPO products (B. Rodamilans, A. Valli, A. Mingot, D. San Leon, D. B. Baulcombe, J. J. Lopez-Moya, and J.A. Garcia, J Virol 89:6965-6967, 2015, doi:10.1128/JVI.00337-15). Here, we demonstrate by liquid chromatography coupled to mass spectrometry that both P1 and P1N-PISPO are produced during viral infection and coexist in SPFMV-infected Ipomoea batatas plants. Interestingly, transient expression of SPFMV gene products coagroinfiltrated with a reporter gene in Nicotiana benthamiana revealed that P1N-PISPO acts as an RNA silencing suppressor, a role normally associated with HCPro in other potyviruses. Moreover, mutation of WG/GW motifs present in P1N-PISPO abolished its silencing suppression activity, suggesting that the function might require interaction with Argonaute components of the silencing machinery, as was shown for other viral suppressors. Altogether, our results reveal a further layer of complexity of the RNA silencing suppression activity within the Potyviridae family.
Importance: Gene products of potyviruses include P1, HCPro, P3, 6K1, CI, 6K2, VPg/NIaPro, NIb, and CP, all derived from the proteolytic processing of a large polyprotein, and an additional P3N-PIPO product, with the PIPO segment encoded in a different frame within the P3 cistron. In sweet potato feathery mottle virus (SPFMV), another out-of-frame element (PISPO) was predicted within the P1 region. We have shown recently that a polymerase slippage mechanism can generate the transcript variants with extra nucleotides that could be translated into P1N-PISPO and P3N-PIPO. Now, we demonstrate by mass spectrometry analysis that P1N-PISPO is indeed produced in SPFMV-infected plants, in addition to P1. Interestingly, while in other potyviruses the suppressor of RNA silencing is HCPro, we show here that P1N-PISPO exhibited this activity in SPFMV, revealing how the complexity of the gene content could contribute to supply this essential function in members of the Potyviridae family.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Valli A, García JA, López-Moya JJ. 2015. Potyviridae, p 1–10. In Encyclopedia of life sciences. John Wiley & Sons, Ltd., Chichester, United Kingdom.
-
- Sakai J, Mori M, Morishita T, Tanaka M, Hanada K, Usugi T, Nishiguchi M. 1997. Complete nucleotide sequence and genome organization of sweet potato feathery mottle virus (S strain) genomic RNA: the large coding region of the P1 gene. Arch Virol 142:1553–1562. doi:10.1007/s007050050179. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

