Cutting Edge: Eomesodermin Is Sufficient To Direct Type 1 Innate Lymphocyte Development into the Conventional NK Lineage
- PMID: 26792802
- PMCID: PMC4744497
- DOI: 10.4049/jimmunol.1502396
Cutting Edge: Eomesodermin Is Sufficient To Direct Type 1 Innate Lymphocyte Development into the Conventional NK Lineage
Abstract
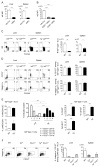
Type 1 innate lymphocytes comprise two developmentally divergent lineages, type 1 helper innate lymphoid cells (hILC1s) and conventional NK cells (cNKs). All type 1 innate lymphocytes (ILCs) express the transcription factor T-bet, but cNKs additionally express Eomesodermin (Eomes). We show that deletion of Eomes alleles at the onset of type 1 ILC maturation using NKp46-Cre imposes a substantial block in cNK development. Formation of the entire lymphoid and nonlymphoid type 1 ILC compartment appears to require the semiredundant action of both T-bet and Eomes. To determine if Eomes is sufficient to redirect hILC1 development to a cNK fate, we generated transgenic mice that express Eomes when and where T-bet is expressed using Tbx21 locus control to drive expression of Eomes codons. Ectopic Eomes induces cNK-like properties across the lymphoid and nonlymphoid type 1 ILC compartments. Subsequent to their divergent lineage specification, hILC1s and cNKs thus possess substantial developmental plasticity.
Copyright © 2016 by The American Association of Immunologists, Inc.
Conflict of interest statement
The authors have no financial conflicts of interest.
Figures


References
-
- Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, Bienvenu J, Henry T, Debien E, Hasan UA, Marvel J, Yoh K, Takahashi S, Prinz I, de Bernard S, Buffat L, Walzer T. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med. 2014;211:563–577. - PMC - PubMed
-
- Cortez VS, Fuchs A, Cella M, Gilfillan S, Colonna M. Cutting edge: Salivary gland NK cells develop independently of Nfil3 in steady-state. J Immunol. 2014;192:4487–4491. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

