Phosphoproteomics Profiling of Tobacco Mature Pollen and Pollen Activated in vitro
- PMID: 26792808
- PMCID: PMC4824859
- DOI: 10.1074/mcp.M115.051672
Phosphoproteomics Profiling of Tobacco Mature Pollen and Pollen Activated in vitro
Abstract
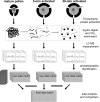
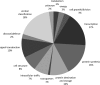
Tobacco mature pollen has extremely desiccated cytoplasm, and is metabolically quiescent. Upon re-hydration it becomes metabolically active and that results in later emergence of rapidly growing pollen tube. These changes in cytoplasm hydration and metabolic activity are accompanied by protein phosphorylation. In this study, we subjected mature pollen, 5-min-activated pollen, and 30-min-activated pollen to TCA/acetone protein extraction, trypsin digestion and phosphopeptide enrichment by titanium dioxide. The enriched fraction was subjected to nLC-MS/MS. We identified 471 phosphopeptides that carried 432 phosphorylation sites, position of which was exactly matched by mass spectrometry. These 471 phosphopeptides were assigned to 301 phosphoproteins, because some proteins carried more phosphorylation sites. Of the 13 functional groups, the majority of proteins were put into these categories: transcription, protein synthesis, protein destination and storage, and signal transduction. Many proteins were of unknown function, reflecting the fact that male gametophyte contains many specific proteins that have not been fully functionally annotated. The quantitative data highlighted the dynamics of protein phosphorylation during pollen activation; the identified phosphopeptides were divided into seven groups based on the regulatory trends. The major group comprised mature pollen-specific phosphopeptides that were dephosphorylated during pollen activation. Several phosphopeptides representing the same phosphoprotein had different regulation, which pinpointed the complexity of protein phosphorylation and its clear functional context. Collectively, we showed the first phosphoproteomics data on activated pollen where the position of phosphorylation sites was clearly demonstrated and regulatory kinetics was resolved.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Mishra N. S., Tuteja R., and Tuteja N. (2006) Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 452, 55–68 - PubMed
-
- Francis D., and Halford N. G. (1995) The plant cell cycle. Physiol. Plant. 93, 365–374
-
- van der Kelen K., Beyaert R., Inze D., and de Veylder L. (2009) Translational control of eukaryotic gene expression. Crit. Rev. Biochem. Mol. Biol. 44, 143–168 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

