A robust platform for chemical genomics in bacterial systems
- PMID: 26792836
- PMCID: PMC4791123
- DOI: 10.1091/mbc.E15-08-0573
A robust platform for chemical genomics in bacterial systems
Abstract
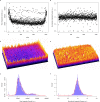
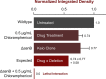
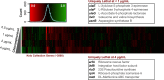
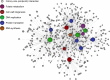
While genetic perturbation has been the conventional route to probing bacterial systems, small molecules are showing great promise as probes for cellular complexity. Indeed, systematic investigations of chemical-genetic interactions can provide new insights into cell networks and are often starting points for understanding the mechanism of action of novel chemical probes. We have developed a robust and sensitive platform for chemical-genomic investigations in bacteria. The approach monitors colony volume kinetically using transmissive scanning measurements, enabling acquisition of growth rates and conventional endpoint measurements. We found that chemical-genomic profiles were highly sensitive to concentration, necessitating careful selection of compound concentrations. Roughly 20,000,000 data points were collected for 15 different antibiotics. While 1052 chemical-genetic interactions were identified using the conventional endpoint biomass approach, adding interactions in growth rate resulted in 1564 interactions, a 50-200% increase depending on the drug, with many genes uncharacterized or poorly annotated. The chemical-genetic interaction maps generated from these data reveal common genes likely involved in multidrug resistance. Additionally, the maps identified deletion backgrounds exhibiting class-specific potentiation, revealing conceivable targets for combination approaches to drug discovery. This open platform is highly amenable to kinetic screening of any arrayable strain collection, be it prokaryotic or eukaryotic.
© 2016 French et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures


References
-
- Bianchi AA, Baneyx F. Hyperosmotic shock induces the sigma32 and sigmaE stress regulons of Escherichia coli. Mol Microbiol. 1999;34:1029–1038. - PubMed
-
- Brideau C, Gunter B, Pikounis B, Liaw A. Improved statistical methods for hit selection in high-throughput screening. J Biomol Screen. 2003;8:634–647. - PubMed
-
- Brochado AR, Typas A. High-throughput approaches to understanding gene function and mapping network architecture in bacteria. Curr Opin Microbiol. 2013;16:199–206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

