Re-evaluation of G-quadruplex propensity with G4Hunter
- PMID: 26792894
- PMCID: PMC4770238
- DOI: 10.1093/nar/gkw006
Re-evaluation of G-quadruplex propensity with G4Hunter
Abstract
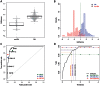
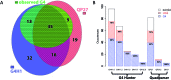
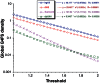
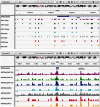
Critical evidence for the biological relevance of G-quadruplexes (G4) has recently been obtained in seminal studies performed in a variety of organisms. Four-stranded G-quadruplex DNA structures are promising drug targets as these non-canonical structures appear to be involved in a number of key biological processes. Given the growing interest for G4, accurate tools to predict G-quadruplex propensity of a given DNA or RNA sequence are needed. Several algorithms such as Quadparser predict quadruplex forming propensity. However, a number of studies have established that sequences that are not detected by these tools do form G4 structures (false negatives) and that other sequences predicted to form G4 structures do not (false positives). Here we report development and testing of a radically different algorithm, G4Hunter that takes into account G-richness and G-skewness of a given sequence and gives a quadruplex propensity score as output. To validate this model, we tested it on a large dataset of 392 published sequences and experimentally evaluated quadruplex forming potential of 209 sequences using a combination of biophysical methods to assess quadruplex formation in vitro. We experimentally validated the G4Hunter algorithm on a short complete genome, that of the human mitochondria (16.6 kb), because of its relatively high GC content and GC skewness as well as the biological relevance of these quadruplexes near instability hotspots. We then applied the algorithm to genomes of a number of species, including humans, allowing us to conclude that the number of sequences capable of forming stable quadruplexes (at least in vitro) in the human genome is significantly higher, by a factor of 2-10, than previously thought.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
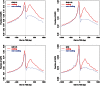
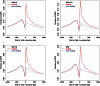
References
-
- Watson J.D., Crick F.H. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. - PubMed
-
- De Cian A., Lacroix L., Douarre C., Temime-Smaali N., Trentesaux C., Riou J.F., Mergny J.L. Targeting telomeres and telomerase. Biochimie. 2008;90:131–155. - PubMed
-
- Wieland M., Hartig J.S. RNA quadruplex-based modulation of gene expression. Chem. Biol. 2007;14:757–763. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

