Histones and histone modifications in perinuclear chromatin anchoring: from yeast to man
- PMID: 26792937
- PMCID: PMC4783997
- DOI: 10.15252/embr.201541809
Histones and histone modifications in perinuclear chromatin anchoring: from yeast to man
Abstract
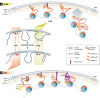
It is striking that within a eukaryotic nucleus, the genome can assume specific spatiotemporal distributions that correlate with the cell's functional states. Cell identity itself is determined by distinct sets of genes that are expressed at a given time. On the level of the individual gene, there is a strong correlation between transcriptional activity and associated histone modifications. Histone modifications act by influencing the recruitment of non-histone proteins and by determining the level of chromatin compaction, transcription factor binding, and transcription elongation. Accumulating evidence also shows that the subnuclear position of a gene or domain correlates with its expression status. Thus, the question arises whether this spatial organization results from or determines a gene's chromatin status. Although the association of a promoter with the inner nuclear membrane (INM) is neither necessary nor sufficient for repression, the perinuclear sequestration of heterochromatin is nonetheless conserved from yeast to man. How does subnuclear localization influence gene expression? Recent work argues that the common denominator between genome organization and gene expression is the modification of histones and in some cases of histone variants. This provides an important link between local chromatin structure and long-range genome organization in interphase cells. In this review, we will evaluate how histones contribute to the latter, and discuss how this might help to regulate genes crucial for cell differentiation.
Keywords: CEC‐4; histone methylation; inner nuclear membrane; nuclear envelope; nuclear organization.
© 2016 The Authors. Published under the terms of the CC BY NC ND 4.0 license.
Figures



References
-
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H (2002) Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296: 158–162 - PubMed
-
- Brown KE, Amoils S, Horn JM, Buckle VJ, Higgs DR, Merkenschlager M, Fisher AG (2001) Expression of alpha‐ and beta‐globin genes occurs within different nuclear domains in haemopoietic cells. Nat Cell Biol 3: 602–606 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

