Milk Bottom-Up Proteomics: Method Optimization
- PMID: 26793233
- PMCID: PMC4707256
- DOI: 10.3389/fgene.2015.00360
Milk Bottom-Up Proteomics: Method Optimization
Abstract
Milk is a complex fluid whose proteome displays a diverse set of proteins of high abundance such as caseins and medium to low abundance whey proteins such as ß-lactoglobulin, lactoferrin, immunoglobulins, glycoproteins, peptide hormones, and enzymes. A sample preparation method that enables high reproducibility and throughput is key in reliably identifying proteins present or proteins responding to conditions such as a diet, health or genetics. Using skim milk samples from Jersey and Holstein-Friesian cows, we compared three extraction procedures which have not previously been applied to samples of cows' milk. Method A (urea) involved a simple dilution of the milk in a urea-based buffer, method B (TCA/acetone) involved a trichloroacetic acid (TCA)/acetone precipitation, and method C (methanol/chloroform) involved a tri-phasic partition method in chloroform/methanol solution. Protein assays, SDS-PAGE profiling, and trypsin digestion followed by nanoHPLC-electrospray ionization-tandem mass spectrometry (nLC-ESI-MS/MS) analyses were performed to assess their efficiency. Replicates were used at each analytical step (extraction, digestion, injection) to assess reproducibility. Mass spectrometry (MS) data are available via ProteomeXchange with identifier PXD002529. Overall 186 unique accessions, major and minor proteins, were identified with a combination of methods. Method C (methanol/chloroform) yielded the best resolved SDS-patterns and highest protein recovery rates, method A (urea) yielded the greatest number of accessions, and, of the three procedures, method B (TCA/acetone) was the least compatible of all with a wide range of downstream analytical procedures. Our results also highlighted breed differences between the proteins in milk of Jersey and Holstein-Friesian cows.
Keywords: Jersey and Holstein-Friesian cow milk; proteome; replicates; shotgun nLC-ESI-MS; trypsin digestion.
Figures
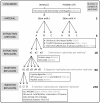


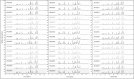

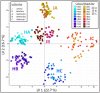


References
-
- Bligh E. G., Dyer W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

