Assembly, Assessment, and Availability of De novo Generated Eukaryotic Transcriptomes
- PMID: 26793234
- PMCID: PMC4707302
- DOI: 10.3389/fgene.2015.00361
Assembly, Assessment, and Availability of De novo Generated Eukaryotic Transcriptomes
Abstract
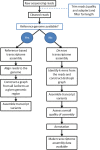
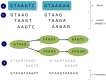
De novo assembly of a complete transcriptome without the need for a guiding reference genome is attractive, particularly where the cost and complexity of generating a eukaryote genome is prohibitive. The transcriptome should not however be seen as just a quick and cheap alternative to building a complete genome. Transcriptomics allows the understanding and comparison of spatial and temporal samples within an organism, and allows surveying of multiple individuals or closely related species. De novo assembly in theory allows the building of a complete transcriptome without any prior knowledge of the genome. It also allows the discovery of alternate splice forms of coding RNAs and also non-coding RNAs, which are often missed by proteomic approaches, or are incompletely annotated in genome studies. The limitations of the method are that the generation of a truly complete assembly is unlikely, and so we require some methods for the assessment of the quality and appropriateness of a generated transcriptome. Whilst no single consensus pipeline or tool is agreed as optimal, various algorithms, and easy to use software do exist making transcriptome generation a more common approach. With this expansion of data, questions still exist relating to how do we make these datasets fully discoverable, comparable and most useful to understand complex biological systems?
Keywords: annotation; assessment; availability; de novo transcriptome assembly; high-throughput sequencing.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

