An efficient approach to BAC based assembly of complex genomes
- PMID: 26793268
- PMCID: PMC4719536
- DOI: 10.1186/s13007-016-0107-9
An efficient approach to BAC based assembly of complex genomes
Abstract
Background: There has been an exponential growth in the number of genome sequencing projects since the introduction of next generation DNA sequencing technologies. Genome projects have increasingly involved assembly of whole genome data which produces inferior assemblies compared to traditional Sanger sequencing of genomic fragments cloned into bacterial artificial chromosomes (BACs). While whole genome shotgun sequencing using next generation sequencing (NGS) is relatively fast and inexpensive, this method is extremely challenging for highly complex genomes, where polyploidy or high repeat content confounds accurate assembly, or where a highly accurate 'gold' reference is required. Several attempts have been made to improve genome sequencing approaches by incorporating NGS methods, to variable success.
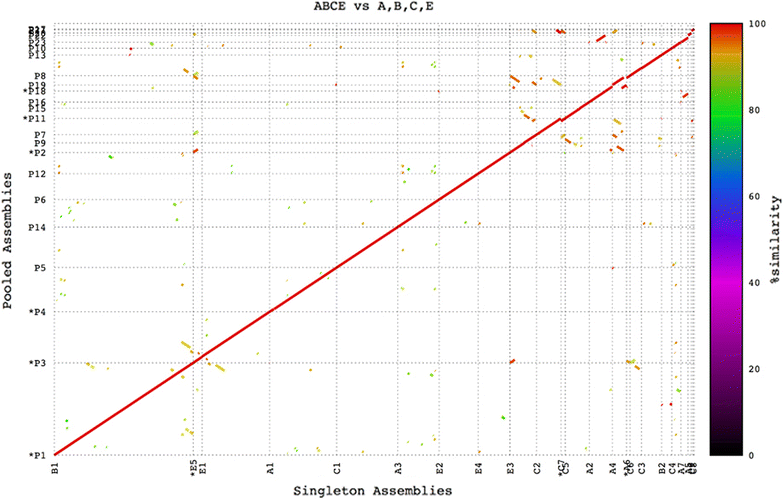
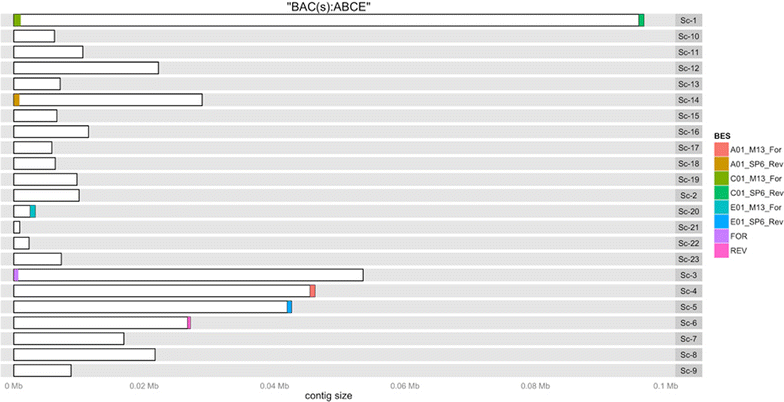
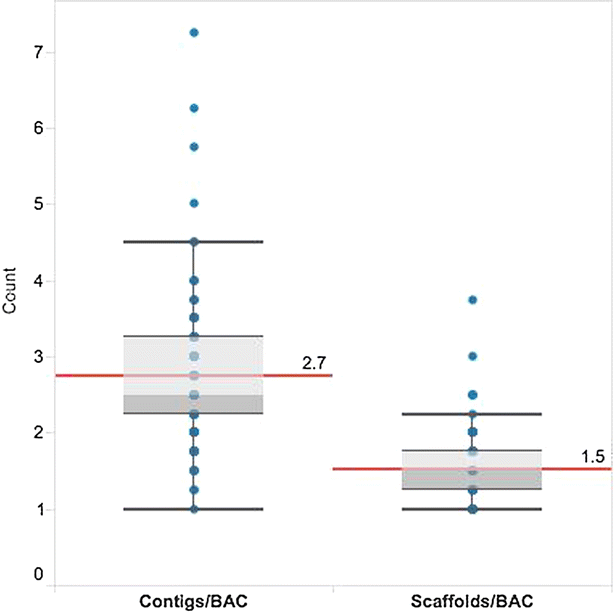
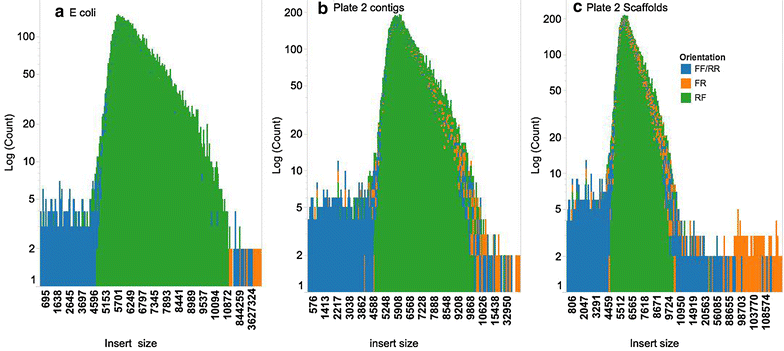
Results: We present the application of a novel BAC sequencing approach which combines indexed pools of BACs, Illumina paired read sequencing, a sequence assembler specifically designed for complex BAC assembly, and a custom bioinformatics pipeline. We demonstrate this method by sequencing and assembling BAC cloned fragments from bread wheat and sugarcane genomes.
Conclusions: We demonstrate that our assembly approach is accurate, robust, cost effective and scalable, with applications for complete genome sequencing in large and complex genomes.
Keywords: 7DS; Assembly; BAC; Next-generation sequencing; SASSY; Saccharum spp; Triticum aestivum.
Figures

References
-
- Ruperao P, Chan C-KK, Azam S, Karafiátová M, Hayashi S, Čížková J, Saxena RK, Šimková H, Song C, Vrána J, Chitikineni A, Visendi P, Gaur PM, Millán T, Singh KB, Tara B, Wang J, Batley J, Doležel J, Varshney RK, Edwards D. A chromosomal genomics approach to assess and validate the desi and kabuli draft chickpea genome assemblies. Plant Biotechnol J. 2014;12:778–786. doi: 10.1111/pbi.12182. - DOI - PubMed
-
- Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, Kohara Y, Fujiyama A, Hayashi T, Itoh T. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24:1384–1395. doi: 10.1101/gr.170720.113. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

