The Arabidopsis glutamyl-tRNA reductase (GluTR) forms a ternary complex with FLU and GluTR-binding protein
- PMID: 26794057
- PMCID: PMC4726326
- DOI: 10.1038/srep19756
The Arabidopsis glutamyl-tRNA reductase (GluTR) forms a ternary complex with FLU and GluTR-binding protein
Abstract
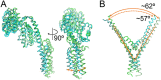
Tetrapyrrole biosynthesis is an essential and tightly regulated process, and glutamyl-tRNA reductase (GluTR) is a key target for multiple regulatory factors at the post-translational level. By binding to the thylakoid membrane protein FLUORESCENT (FLU) or the soluble stromal GluTR-binding protein (GBP), the activity of GluTR is down- or up-regulated. Here, we reconstructed a ternary complex composed of the C-terminal tetratricopepetide-repeat domain of FLU, GBP, and GluTR, crystallized and solved the structure of the complex at 3.2 Å. The overall structure resembles the shape of merged two binary complexes as previously reported, and shows a large conformational change within GluTR. We also demonstrated that GluTR binds tightly with GBP but does not bind to GSAM under the same condition. These findings allow us to suggest a biological role of the ternary complex for the regulation of plant GluTR.
Figures


Similar articles
-
The Non-canonical Tetratricopeptide Repeat (TPR) Domain of Fluorescent (FLU) Mediates Complex Formation with Glutamyl-tRNA Reductase.J Biol Chem. 2015 Jul 10;290(28):17559-65. doi: 10.1074/jbc.M115.662981. Epub 2015 Jun 2. J Biol Chem. 2015. PMID: 26037924 Free PMC article.
-
Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein.Proc Natl Acad Sci U S A. 2014 May 6;111(18):6630-5. doi: 10.1073/pnas.1400166111. Epub 2014 Apr 21. Proc Natl Acad Sci U S A. 2014. PMID: 24753615 Free PMC article.
-
The GluTR-binding protein is the heme-binding factor for feedback control of glutamyl-tRNA reductase.Elife. 2019 Jun 13;8:e46300. doi: 10.7554/eLife.46300. Elife. 2019. PMID: 31194674 Free PMC article.
-
Posttranslational Control of ALA Synthesis Includes GluTR Degradation by Clp Protease and Stabilization by GluTR-Binding Protein.Plant Physiol. 2016 Apr;170(4):2040-51. doi: 10.1104/pp.15.01945. Epub 2016 Feb 16. Plant Physiol. 2016. PMID: 26884485 Free PMC article.
-
New insights in the topology of the biosynthesis of 5-aminolevulinic acid.Plant Signal Behav. 2013 Feb;8(2):e23124. doi: 10.4161/psb.23124. Epub 2013 Jan 8. Plant Signal Behav. 2013. PMID: 23299429 Free PMC article. Review.
Cited by
-
Regulatory and retrograde signaling networks in the chlorophyll biosynthetic pathway.J Integr Plant Biol. 2025 Apr;67(4):887-911. doi: 10.1111/jipb.13837. Epub 2025 Jan 24. J Integr Plant Biol. 2025. PMID: 39853950 Free PMC article. Review.
-
Effect of chlorophyll biosynthesis-related genes on the leaf color in Hosta (Hosta plantaginea Aschers) and tobacco (Nicotiana tabacum L.).BMC Plant Biol. 2021 Jan 15;21(1):45. doi: 10.1186/s12870-020-02805-6. BMC Plant Biol. 2021. PMID: 33451287 Free PMC article.
-
Cold acclimation can specifically inhibit chlorophyll biosynthesis in young leaves of Pakchoi.BMC Plant Biol. 2021 Apr 10;21(1):172. doi: 10.1186/s12870-021-02954-2. BMC Plant Biol. 2021. PMID: 33838654 Free PMC article.
-
A novel tetratricopeptide-repeat protein, TTP1, forms complexes with glutamyl-tRNA reductase and protochlorophyllide oxidoreductase during tetrapyrrole biosynthesis.J Exp Bot. 2024 Mar 27;75(7):2027-2045. doi: 10.1093/jxb/erad491. J Exp Bot. 2024. PMID: 38070484 Free PMC article.
-
Chloroplast SRP43 acts as a chaperone for glutamyl-tRNA reductase, the rate-limiting enzyme in tetrapyrrole biosynthesis.Proc Natl Acad Sci U S A. 2018 Apr 10;115(15):E3588-E3596. doi: 10.1073/pnas.1719645115. Epub 2018 Mar 26. Proc Natl Acad Sci U S A. 2018. PMID: 29581280 Free PMC article.
References
-
- Tanaka R. & Tanaka A. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346 (2007). - PubMed
-
- Mochizuki N. et al.. The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci. 15, 488–498 (2010). - PubMed
-
- Vothknecht U. C., Kannangara C. G. & von Wettstein D. Barley glutamyl tRNAGlu reductase: Mutations affecting haem inhibition and enzyme activity. Phytochemistry 47, 513–519 (1998). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

