Cross-kingdom inhibition of breast cancer growth by plant miR159
- PMID: 26794868
- PMCID: PMC4746606
- DOI: 10.1038/cr.2016.13
Cross-kingdom inhibition of breast cancer growth by plant miR159
Abstract
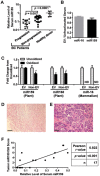
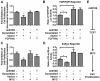
MicroRNAs (miRNAs) are critical regulators of gene expression, and exert extensive impacts on development, physiology, and disease of eukaryotes. A high degree of parallelism is found in the molecular basis of miRNA biogenesis and action in plants and animals. Recent studies interestingly suggest a potential cross-kingdom action of plant-derived miRNAs, through dietary intake, in regulating mammalian gene expression. Although the source and scope of plant miRNAs detected in mammalian specimens remain controversial, these initial studies inspired us to determine whether plant miRNAs can be detected in Western human sera and whether these plant miRNAs are able to influence gene expression and cellular processes related to human diseases such as cancer. Here we found that Western donor sera contained the plant miRNA miR159, whose abundance in the serum was inversely correlated with breast cancer incidence and progression in patients. In human sera, miR159 was predominantly detected in the extracellular vesicles, and was resistant to sodium periodate oxidation suggesting the plant-originated 2'-O-methylation on the 3' terminal ribose. In breast cancer cells but not non-cancerous mammary epithelial cells, a synthetic mimic of miR159 was capable of inhibiting proliferation by targeting TCF7 that encodes a Wnt signaling transcription factor, leading to a decrease in MYC protein levels. Oral administration of miR159 mimic significantly suppressed the growth of xenograft breast tumors in mice. These results demonstrate for the first time that a plant miRNA can inhibit cancer growth in mammals.
Figures




References
-
- 1Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–297. - PubMed
-
- 3Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6:857–866. - PubMed
-
- 4Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65:7065–7070. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

