Skin Keratins
- PMID: 26795476
- PMCID: PMC4902878
- DOI: 10.1016/bs.mie.2015.09.032
Skin Keratins
Abstract
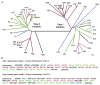
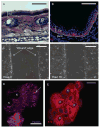
Keratins comprise the type I and type II intermediate filament-forming proteins and occur primarily in epithelial cells. They are encoded by 54 evolutionarily conserved genes (28 type I, 26 type II) and regulated in a pairwise and tissue type-, differentiation-, and context-dependent manner. Keratins serve multiple homeostatic and stress-enhanced mechanical and nonmechanical functions in epithelia, including the maintenance of cellular integrity, regulation of cell growth and migration, and protection from apoptosis. These functions are tightly regulated by posttranslational modifications as well as keratin-associated proteins. Genetically determined alterations in keratin-coding sequences underlie highly penetrant and rare disorders whose pathophysiology reflects cell fragility and/or altered tissue homeostasis. Moreover, keratin mutation or misregulation represents risk factors or genetic modifiers for several acute and chronic diseases. This chapter focuses on keratins that are expressed in skin epithelia, and details a number of basic protocols and assays that have proven useful for analyses being carried out in skin.
Keywords: Cytoskeleton; Differentiation; Epidermis; Genetic disease; Intermediate filament; Keratin; Keratinocyte; Primary cell culture; Skin.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


References
-
- Bernot KM, Coulombe PA, McGowan KM. Keratin 16 expression defines a subset of epithelial cells during skin morphogenesis and the hair cycle. The Journal of Investigative Dermatology. 2002;119:1137–1149. - PubMed
-
- Bernot KM, Coulombe PA, Wong P. Skin: An ideal model system to study keratin genes and proteins. Methods in Cell Biology. 2004;78:453–487. - PubMed
-
- Bonifas JM, Rothman AL, Epstein EH., Jr Epidermolysis bullosa simplex: Evidence in two families for keratin gene abnormalities. Science. 1991;254:1202–1205. - PubMed
-
- Byrne C. Regulation of gene expression in developing epidermal epithelia. Bioessays. 1997;19:691–698. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

