CRISPR-Cas9 Can Inhibit HIV-1 Replication but NHEJ Repair Facilitates Virus Escape
- PMID: 26796669
- PMCID: PMC4786927
- DOI: 10.1038/mt.2016.24
CRISPR-Cas9 Can Inhibit HIV-1 Replication but NHEJ Repair Facilitates Virus Escape
Abstract
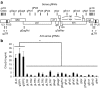
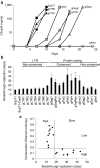
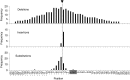
Several recent studies demonstrated that the clustered regularly interspaced short palindromic repeats (CRISPR)-associated endonuclease Cas9 can be used for guide RNA (gRNA)-directed, sequence-specific cleavage of HIV proviral DNA in infected cells. We here demonstrate profound inhibition of HIV-1 replication by harnessing T cells with Cas9 and antiviral gRNAs. However, the virus rapidly and consistently escaped from this inhibition. Sequencing of the HIV-1 escape variants revealed nucleotide insertions, deletions, and substitutions around the Cas9/gRNA cleavage site that are typical for DNA repair by the nonhomologous end-joining pathway. We thus demonstrate the potency of CRISPR-Cas9 as an antiviral approach, but any therapeutic strategy should consider the viral escape implications.
Figures

Comment in
-
Recent advances in CRISPR/Cas9 technology for targeting latent HIV-1 reservoirs.AIDS. 2016 Jul 17;30(11):N17. doi: 10.1097/QAD.0000000000001138. AIDS. 2016. PMID: 27139312 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

