In Vivo Analysis of Disease-Associated Point Mutations Unveils Profound Differences in mRNA Splicing of Peripherin-2 in Rod and Cone Photoreceptors
- PMID: 26796962
- PMCID: PMC4722987
- DOI: 10.1371/journal.pgen.1005811
In Vivo Analysis of Disease-Associated Point Mutations Unveils Profound Differences in mRNA Splicing of Peripherin-2 in Rod and Cone Photoreceptors
Abstract
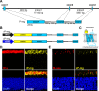
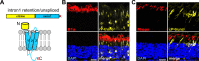
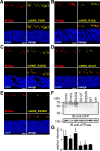
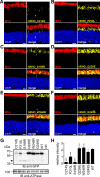
Point mutations in peripherin-2 (PRPH2) are associated with severe retinal degenerative disorders affecting rod and/or cone photoreceptors. Various disease-causing mutations have been identified, but the exact contribution of a given mutation to the clinical phenotype remains unclear. Exonic point mutations are usually assumed to alter single amino acids, thereby influencing specific protein characteristics; however, they can also affect mRNA splicing. To examine the effects of distinct PRPH2 point mutations on mRNA splicing and protein expression in vivo, we designed PRPH2 minigenes containing the three coding exons and relevant intronic regions of human PRPH2. Minigenes carrying wild type PRPH2 or PRPH2 exon 2 mutations associated with rod or cone disorders were expressed in murine photoreceptors using recombinant adeno-associated virus (rAAV) vectors. We detect three PRPH2 splice isoforms in rods and cones: correctly spliced, intron 1 retention, and unspliced. In addition, we show that only the correctly spliced isoform results in detectable protein expression. Surprisingly, compared to rods, differential splicing leads to lower expression of correctly spliced and higher expression of unspliced PRPH2 in cones. These results were confirmed in qRT-PCR experiments from FAC-sorted murine rods and cones. Strikingly, three out of five cone disease-causing PRPH2 mutations profoundly enhanced correct splicing of PRPH2, which correlated with strong upregulation of mutant PRPH2 protein expression in cones. By contrast, four out of six PRPH2 mutants associated with rod disorders gave rise to a reduced PRPH2 protein expression via different mechanisms. These mechanisms include aberrant mRNA splicing, protein mislocalization, and protein degradation. Our data suggest that upregulation of PRPH2 levels in combination with defects in the PRPH2 function caused by the mutation might be an important mechanism leading to cone degeneration. By contrast, the pathology of rod-specific PRPH2 mutations is rather characterized by PRPH2 downregulation and impaired protein localization.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Goldberg AF. Role of peripherin/rds in vertebrate photoreceptor architecture and inherited retinal degenerations. Int Rev Cytol. 2006;253: 131–175. - PubMed
-
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8(10): 749–761. - PubMed
-
- Becirovic E, Ebermann I, Nagy D, Zrenner E, Seeliger MW, Bolz HJ. Usher syndrome type 1 due to missense mutations on both CDH23 alleles: investigation of mRNA splicing. Hum Mutat. 2008;29(3): 452. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

