Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells
- PMID: 26797145
- PMCID: PMC4811353
- DOI: 10.1126/science.aad5510
Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells
Abstract
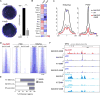
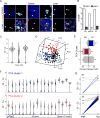
Differentiated macrophages can self-renew in tissues and expand long term in culture, but the gene regulatory mechanisms that accomplish self-renewal in the differentiated state have remained unknown. Here we show that in mice, the transcription factors MafB and c-Maf repress a macrophage-specific enhancer repertoire associated with a gene network that controls self-renewal. Single-cell analysis revealed that, in vivo, proliferating resident macrophages can access this network by transient down-regulation of Maf transcription factors. The network also controls embryonic stem cell self-renewal but is associated with distinct embryonic stem cell-specific enhancers. This indicates that distinct lineage-specific enhancer platforms regulate a shared network of genes that control self-renewal potential in both stem and mature cells.
Copyright © 2016, American Association for the Advancement of Science.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

