Integrative DNA methylome analysis of pan-cancer biomarkers in cancer discordant monozygotic twin-pairs
- PMID: 26798410
- PMCID: PMC4721070
- DOI: 10.1186/s13148-016-0172-y
Integrative DNA methylome analysis of pan-cancer biomarkers in cancer discordant monozygotic twin-pairs
Abstract
Background: A key focus in cancer research is the discovery of biomarkers that accurately diagnose early lesions in non-invasive tissues. Several studies have identified malignancy-associated DNA methylation changes in blood, yet no general cancer biomarker has been identified to date. Here, we explore the potential of blood DNA methylation as a biomarker of pan-cancer (cancer of multiple different origins) in 41 female cancer discordant monozygotic (MZ) twin-pairs sampled before or after diagnosis using the Illumina HumanMethylation450 BeadChip.
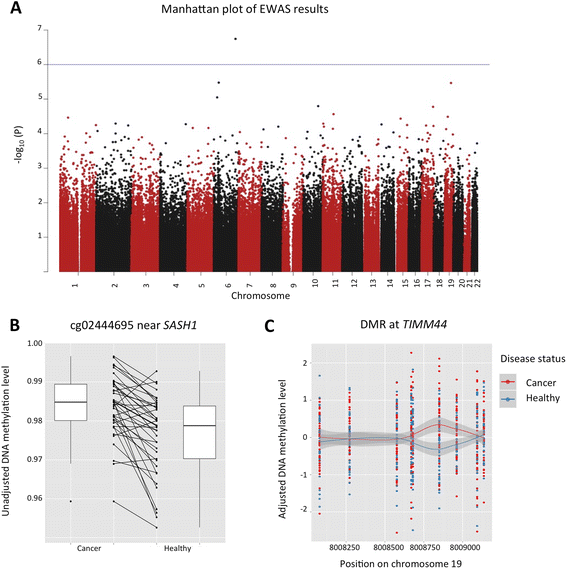
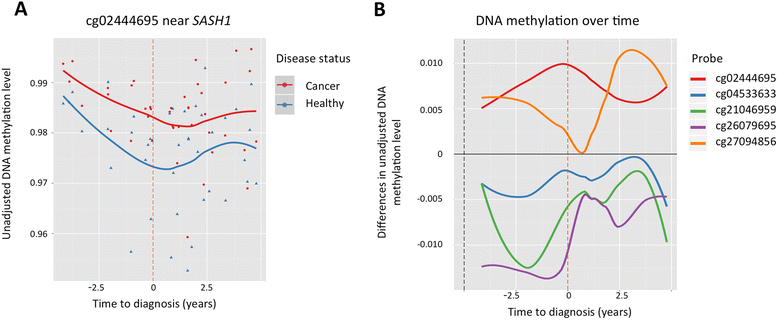
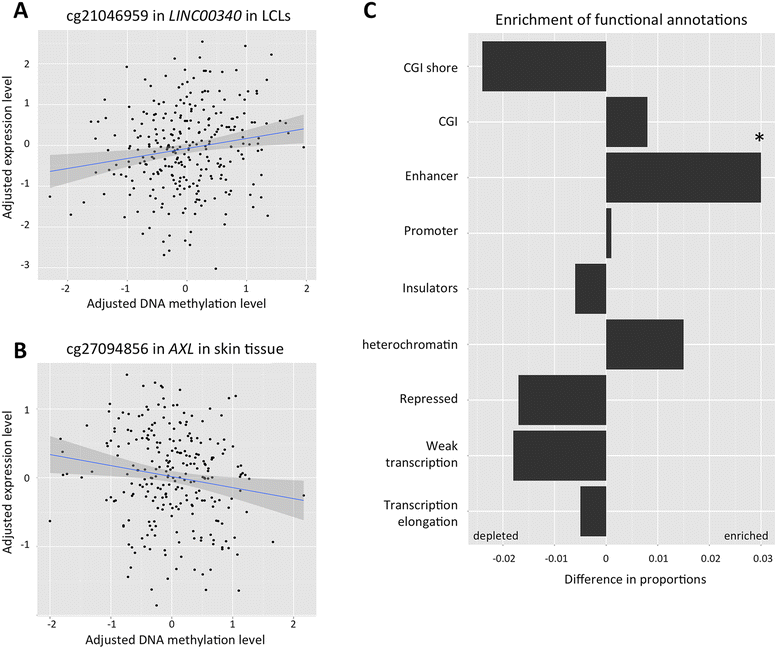
Results: We analysed epigenome-wide DNA methylation profiles in 41 cancer discordant MZ twin-pairs with affected individuals diagnosed with tumours at different single primary sites: the breast, cervix, colon, endometrium, thyroid gland, skin (melanoma), ovary, and pancreas. No significant global differences in whole blood DNA methylation profiles were observed. Epigenome-wide analyses identified one novel pan-cancer differentially methylated position at false discovery rate (FDR) threshold of 10 % (cg02444695, P = 1.8 × 10(-7)) in an intergenic region 70 kb upstream of the SASH1 tumour suppressor gene, and three suggestive signals in COL11A2, AXL, and LINC00340. Replication of the four top-ranked signals in an independent sample of nine cancer-discordant MZ twin-pairs showed a similar direction of association at COL11A2, AXL, and LINC00340, and significantly greater methylation discordance at AXL compared to 480 healthy concordant MZ twin-pairs. The effects at cg02444695 (near SASH1), COL11A2, and LINC00340 were the most promising in biomarker potential because the DNA methylation differences were found to pre-exist in samples obtained prior to diagnosis and were limited to a 5-year period before diagnosis. Gene expression follow-up at the top-ranked signals in 283 healthy individuals showed correlation between blood methylation and gene expression in lymphoblastoid cell lines at PRL, and in the skin tissue at AXL. A significant enrichment of differential DNA methylation was observed in enhancer regions (P = 0.03).
Conclusions: We identified DNA methylation signatures in blood associated with pan-cancer, at or near SASH1, COL11A2, AXL, and LINC00340. Three of these signals were present up to 5 years prior to cancer diagnosis, highlighting the potential clinical utility of whole blood DNA methylation analysis in cancer surveillance.
Keywords: Biomarker; Cancer; DNA methylation; Discordant monozygotic twins; Epigenetics; Twin study.
Figures

Similar articles
-
Epigenome-wide analysis in newborn blood spots from monozygotic twins discordant for cerebral palsy reveals consistent regional differences in DNA methylation.Clin Epigenetics. 2018 Feb 23;10:25. doi: 10.1186/s13148-018-0457-4. eCollection 2018. Clin Epigenetics. 2018. PMID: 29484035 Free PMC article.
-
Effects of smoking on genome-wide DNA methylation profiles: A study of discordant and concordant monozygotic twin pairs.Elife. 2023 Aug 10;12:e83286. doi: 10.7554/eLife.83286. Elife. 2023. PMID: 37643467 Free PMC article.
-
DNA Methylation Changes in the IGF1R Gene in Birth Weight Discordant Adult Monozygotic Twins.Twin Res Hum Genet. 2015 Dec;18(6):635-46. doi: 10.1017/thg.2015.76. Epub 2015 Nov 13. Twin Res Hum Genet. 2015. PMID: 26563994
-
Identical but not the same: the value of discordant monozygotic twins in genetic research.Am J Med Genet B Neuropsychiatr Genet. 2010 Sep;153B(6):1134-49. doi: 10.1002/ajmg.b.31091. Am J Med Genet B Neuropsychiatr Genet. 2010. PMID: 20468073 Review.
-
Using epigenomic studies in monozygotic twins to improve our understanding of cancer.Epigenomics. 2014 Jun;6(3):299-309. doi: 10.2217/epi.14.13. Epigenomics. 2014. PMID: 25111484 Review.
Cited by
-
Value of DNA methylation in predicting curve progression in patients with adolescent idiopathic scoliosis.EBioMedicine. 2018 Oct;36:489-496. doi: 10.1016/j.ebiom.2018.09.014. Epub 2018 Sep 18. EBioMedicine. 2018. PMID: 30241917 Free PMC article.
-
Insulin-like growth factor 2 hypermethylation in peripheral blood leukocytes and colorectal cancer risk and prognosis: a propensity score analysis.Front Oncol. 2023 May 5;13:971435. doi: 10.3389/fonc.2023.971435. eCollection 2023. Front Oncol. 2023. PMID: 37213278 Free PMC article.
-
HMGB1 contributes to SASH1 methylation to attenuate astrocyte adhesion.Cell Death Dis. 2019 May 28;10(6):417. doi: 10.1038/s41419-019-1645-7. Cell Death Dis. 2019. PMID: 31138780 Free PMC article.
-
Developmental hematopoietic stem cell variation explains clonal hematopoiesis later in life.Nat Commun. 2024 Nov 26;15(1):10268. doi: 10.1038/s41467-024-54711-2. Nat Commun. 2024. PMID: 39592593 Free PMC article.
-
Elevated CELSR3 expression is associated with hepatocarcinogenesis and poor prognosis.Oncol Lett. 2020 Aug;20(2):1083-1092. doi: 10.3892/ol.2020.11671. Epub 2020 May 22. Oncol Lett. 2020. PMID: 32724347 Free PMC article.
References
-
- International Agency for Research on Cancer. World Cancer Report 2014. WHO Press; 2014
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

