Topical Review: Molecular reaction and solvation visualized by time-resolved X-ray solution scattering: Structure, dynamics, and their solvent dependence
- PMID: 26798770
- PMCID: PMC4711596
- DOI: 10.1063/1.4865234
Topical Review: Molecular reaction and solvation visualized by time-resolved X-ray solution scattering: Structure, dynamics, and their solvent dependence
Abstract
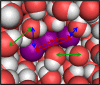
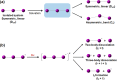
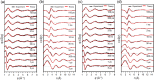
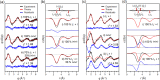
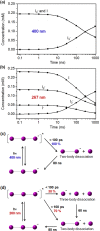
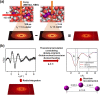
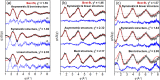
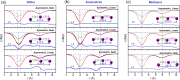
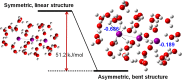
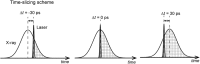
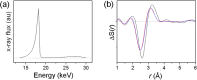
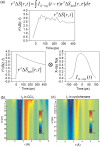
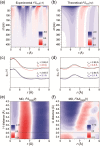
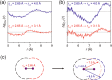
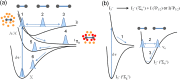
Time-resolved X-ray solution scattering is sensitive to global molecular structure and can track the dynamics of chemical reactions. In this article, we review our recent studies on triiodide ion (I3 (-)) and molecular iodine (I2) in solution. For I3 (-), we elucidated the excitation wavelength-dependent photochemistry and the solvent-dependent ground-state structure. For I2, by combining time-slicing scheme and deconvolution data analysis, we mapped out the progression of geminate recombination and the associated structural change in the solvent cage. With the aid of X-ray free electron lasers, even clearer observation of ultrafast chemical events will be made possible in the near future.
Figures






References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

