Abundant cytomegalovirus (CMV) reactive clonotypes in the CD8(+) T cell receptor alpha repertoire following allogeneic transplantation
- PMID: 26800118
- PMCID: PMC4872374
- DOI: 10.1111/cei.12770
Abundant cytomegalovirus (CMV) reactive clonotypes in the CD8(+) T cell receptor alpha repertoire following allogeneic transplantation
Abstract
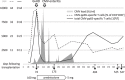
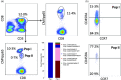
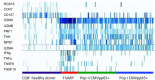
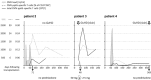
Allogeneic stem cell transplantation is potentially curative, but associated with post-transplantation complications, including cytomegalovirus (CMV) infections. An effective immune response requires T cells recognizing CMV epitopes via their T cell receptors (TCRs). Little is known about the TCR repertoire, in particular the TCR-α repertoire and its clinical relevance in patients following stem cell transplantation. Using next-generation sequencing we examined the TCR-α repertoire of CD8(+) T cells and CMV-specific CD8(+) T cells in four patients. Additionally, we performed single-cell TCR-αβ sequencing of CMV-specific CD8(+) T cells. The TCR-α composition of human leucocyte antigen (HLA)-A*0201 CMVpp65- and CMVIE -specific T cells was oligoclonal and defined by few dominant clonotypes. Frequencies of single clonotypes reached up to 11% of all CD8(+) T cells and half of the total CD8(+) T cell repertoire was dominated by few CMV-reactive clonotypes. Some TCR-α clonotypes were shared between patients. Gene expression of the circulating CMV-specific CD8(+) T cells was consistent with chronically activated effector memory T cells. The CD8(+) T cell response to CMV reactivation resulted in an expansion of a few TCR-α clonotypes to dominate the CD8(+) repertoires. These results warrant further larger studies to define the ability of oligoclonally expanded T cell clones to achieve an effective anti-viral T cell response in this setting.
Keywords: CMV; T cell receptor alpha; T cell receptor repertoire; allogeneic transplantation; next-generation sequencing.
© 2016 British Society for Immunology.
Figures


Similar articles
-
Recognition of distinct cross-reactive virus-specific CD8+ T cells reveals a unique TCR signature in a clinical setting.J Immunol. 2014 Jun 1;192(11):5039-49. doi: 10.4049/jimmunol.1303147. Epub 2014 Apr 28. J Immunol. 2014. PMID: 24778446 Clinical Trial.
-
Cytomegalovirus-Mediated T Cell Receptor Repertoire Perturbation Is Present in Early Life.Front Immunol. 2020 Sep 30;11:1587. doi: 10.3389/fimmu.2020.01587. eCollection 2020. Front Immunol. 2020. PMID: 33101265 Free PMC article.
-
T-cell receptor repertoire of cytomegalovirus-specific cytotoxic T-cells after allogeneic stem cell transplantation.Sci Rep. 2020 Dec 17;10(1):22218. doi: 10.1038/s41598-020-79363-2. Sci Rep. 2020. PMID: 33335252 Free PMC article.
-
Refining human T-cell immunotherapy of cytomegalovirus disease: a mouse model with 'humanized' antigen presentation as a new preclinical study tool.Med Microbiol Immunol. 2016 Dec;205(6):549-561. doi: 10.1007/s00430-016-0471-0. Epub 2016 Aug 18. Med Microbiol Immunol. 2016. PMID: 27539576 Review.
-
Potential Beneficial Effects of Cytomegalovirus Infection after Transplantation.Front Immunol. 2018 Mar 1;9:389. doi: 10.3389/fimmu.2018.00389. eCollection 2018. Front Immunol. 2018. PMID: 29545802 Free PMC article. Review.
Cited by
-
Analysis of the Whole CDR3 T Cell Receptor Repertoire after Hematopoietic Stem Cell Transplantation in 2 Clinical Cohorts.Biol Blood Marrow Transplant. 2020 Jun;26(6):1050-1070. doi: 10.1016/j.bbmt.2020.01.020. Epub 2020 Feb 18. Biol Blood Marrow Transplant. 2020. PMID: 32081787 Free PMC article. Clinical Trial.
-
Sequence and Structural Analyses Reveal Distinct and Highly Diverse Human CD8+ TCR Repertoires to Immunodominant Viral Antigens.Cell Rep. 2017 Apr 18;19(3):569-583. doi: 10.1016/j.celrep.2017.03.072. Cell Rep. 2017. PMID: 28423320 Free PMC article.
-
Mapping the evolution of T cell states during response and resistance to adoptive cellular therapy.Cell Rep. 2021 Nov 9;37(6):109992. doi: 10.1016/j.celrep.2021.109992. Cell Rep. 2021. PMID: 34758319 Free PMC article.
-
Elucidating the Role of the T Cell Receptor Repertoire in Myelodysplastic Neoplasms and Acute Myeloid Leukemia.Diseases. 2025 Jan 17;13(1):19. doi: 10.3390/diseases13010019. Diseases. 2025. PMID: 39851483 Free PMC article. Review.
-
Characterization of human CMV-specific CD8+ T cells using multi-layer single-cell omics.Cell Rep Methods. 2025 Jul 21;5(7):101085. doi: 10.1016/j.crmeth.2025.101085. Epub 2025 Jun 23. Cell Rep Methods. 2025. PMID: 40555232 Free PMC article.
References
-
- Boeckh M. Complications, diagnosis, management, and prevention of CMV infections: current and future. Hematology Am Soc Hematol Educ Program 2011; 2011:305–9. - PubMed
-
- Hebart H, Einsele H. Clinical aspects of CMV infection after stem cell transplantation. Hum Immunol 2004; 65:432–36. - PubMed
-
- Boeckh M, Murphy WJ, Peggs KS. Recent advances in cytomegalovirus: an update on pharmacologic and cellular therapies. Biol Blood Marrow Transplant 2015; 21:24–9. - PubMed
-
- Meier J, Roberts C, Avent K et al Fractal organization of the human T cell repertoire in health and after stem cell transplantation. Biol Blood Marrow Transplant 2013; 19:366–77. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

