Expression of the Long Non-Coding RNA HOTAIR Correlates with Disease Progression in Bladder Cancer and Is Contained in Bladder Cancer Patient Urinary Exosomes
- PMID: 26800519
- PMCID: PMC4723257
- DOI: 10.1371/journal.pone.0147236
Expression of the Long Non-Coding RNA HOTAIR Correlates with Disease Progression in Bladder Cancer and Is Contained in Bladder Cancer Patient Urinary Exosomes
Abstract
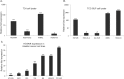
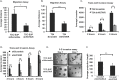
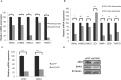
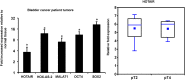
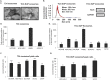
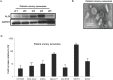
Exosomes are 30-150nM membrane-bound secreted vesicles that are readily isolated from biological fluids such as urine (UEs). Exosomes contain proteins, micro RNA (miRNA), messenger RNA (mRNA), and long non-coding RNA (lncRNA) from their cells of origin. Although miRNA, protein and lncRNA have been isolated from serum as potential biomarkers for benign and malignant disease, it is unknown if lncRNAs in UEs from urothelial bladder cancer (UBC) patients can serve as biomarkers. lncRNAs are > 200 nucleotide long transcripts that do not encode protein and play critical roles in tumor biology. As the number of recognized tumor-associated lncRNAs continues to increase, there is a parallel need to include lncRNAs into biomarker discovery and therapeutic target algorithms. The lncRNA HOX transcript antisense RNA (HOTAIR) has been shown to facilitate tumor initiation and progression and is associated with poor prognosis in several cancers. The importance of HOTAIR in cancer biology has sparked interest in using HOTAIR as a biomarker and potential therapeutic target. Here we show HOTAIR and several tumor-associated lncRNAs are enriched in UEs from UBC patients with high-grade muscle-invasive disease (HGMI pT2-pT4). Knockdown of HOTAIR in UBC cell lines reduces in vitro migration and invasion. Importantly, loss of HOTAIR expression in UBC cell lines alters expression of epithelial-to-mesenchyme transition (EMT) genes including SNAI1, TWIST1, ZEB1, ZO1, MMP1 LAMB3, and LAMC2. Finally, we used RNA-sequencing to identify four additional lncRNAs enriched in UBC patient UEs. These data, suggest that UE-derived lncRNA may potentially serve as biomarkers and therapeutic targets.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

