Genome-wide dissection of AP2/ERF and HSP90 gene families in five legumes and expression profiles in chickpea and pigeonpea
- PMID: 26800652
- PMCID: PMC5066796
- DOI: 10.1111/pbi.12520
Genome-wide dissection of AP2/ERF and HSP90 gene families in five legumes and expression profiles in chickpea and pigeonpea
Abstract
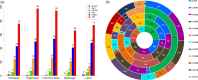
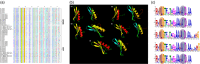
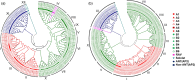
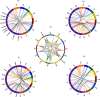
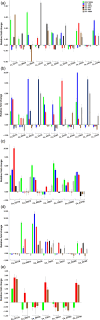
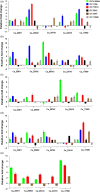
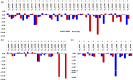
APETALA2/ethylene response factor (AP2/ERF) and heat-shock protein 90 (HSP90) are two significant classes of transcription factor and molecular chaperone proteins which are known to be implicated under abiotic and biotic stresses. Comprehensive survey identified a total of 147 AP2/ERF genes in chickpea, 176 in pigeonpea, 131 in Medicago, 179 in common bean and 140 in Lotus, whereas the number of HSP90 genes ranged from 5 to 7 in five legumes. Sequence alignment and phylogenetic analyses distinguished AP2, ERF, DREB, RAV and soloist proteins, while HSP90 proteins segregated on the basis of their cellular localization. Deeper insights into the gene structure allowed ERF proteins to be classified into AP2s based on DNA-binding domains, intron arrangements and phylogenetic grouping. RNA-seq and quantitative real-time PCR (qRT-PCR) analyses in heat-stressed chickpea as well as Fusarium wilt (FW)- and sterility mosaic disease (SMD)-stressed pigeonpea provided insights into the modus operandi of AP2/ERF and HSP90 genes. This study identified potential candidate genes in response to heat stress in chickpea while for FW and SMD stresses in pigeonpea. For instance, two DREB genes (Ca_02170 and Ca_16631) and three HSP90 genes (Ca_23016, Ca_09743 and Ca_25602) in chickpea can be targeted as potential candidate genes. Similarly, in pigeonpea, a HSP90 gene, C.cajan_27949, was highly responsive to SMD in the resistant genotype ICPL 20096, can be recommended for further functional validation. Also, two DREB genes, C.cajan_41905 and C.cajan_41951, were identified as leads for further investigation in response to FW stress in pigeonpea.
Keywords: AP2/ERF; HSP90; chickpea; pigeonpea.
© 2016 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Alonso, J.M. , Stepanova, A.N. , Leisse, T.J. , Kim, C.J. , Chen, H. , Shinn, P. , Stevenson, D.K. et al. (2003) Genome‐wide insertional mutagenesis of Arabidopsis thaliana . Science, 301, 653–657. - PubMed
-
- Champion, A. , Hebrard, E. , Parra, B. , Bournaud, C. , Marmey, P. , Tranchant, C. and Nicole, M. (2009) Molecular diversity and gene expression of cotton ERF transcription factors reveal that group IXa members are responsive to jasmonate, ethylene and Xanthomonas. Mol. Plant Pathol. 10, 471–485. - PMC - PubMed
-
- Deokar, A.A. , Kondawar, V. , Kohli, D. , Aslam, M. , Jain, P.K. , Karuppayil, S.M. , Varshney, R.K. et al. (2015) The CarERF genes in chickpea (Cicer arietinum L.) and the identification of CarERF116 as abiotic stress responsive transcription factor. Funct. Integr. Genomics, 15, 27–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

