Metabolic regulation of triacylglycerol accumulation in the green algae: identification of potential targets for engineering to improve oil yield
- PMID: 26801206
- PMCID: PMC5066758
- DOI: 10.1111/pbi.12523
Metabolic regulation of triacylglycerol accumulation in the green algae: identification of potential targets for engineering to improve oil yield
Abstract
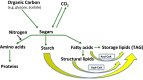
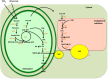
The great need for more sustainable alternatives to fossil fuels has increased our research interests in algal biofuels. Microalgal cells, characterized by high photosynthetic efficiency and rapid cell division, are an excellent source of neutral lipids as potential fuel stocks. Various stress factors, especially nutrient-starvation conditions, induce an increased formation of lipid bodies filled with triacylglycerol in these cells. Here we review our knowledge base on glycerolipid synthesis in the green algae with an emphasis on recent studies on carbon flux, redistribution of lipids under nutrient-limiting conditions and its regulation. We discuss the contributions and limitations of classical and novel approaches used to elucidate the algal triacylglycerol biosynthetic pathway and its regulatory network in green algae. Also discussed are gaps in knowledge and suggestions for much needed research both on the biology of triacylglycerol accumulation and possible avenues to engineer improved algal strains.
Keywords: acyltransferases; algae; biofuel; fatty acids; lipid; lipid droplets; nutrient starvation; transcription factors; triacylglycerol.
© 2016 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures
Similar articles
-
Induction of oil accumulation by heat stress is metabolically distinct from N stress in the green microalgae Coccomyxa subellipsoidea C169.PLoS One. 2018 Sep 27;13(9):e0204505. doi: 10.1371/journal.pone.0204505. eCollection 2018. PLoS One. 2018. PMID: 30261009 Free PMC article.
-
Evaluation of filamentous green algae as feedstocks for biofuel production.Bioresour Technol. 2016 Nov;220:407-413. doi: 10.1016/j.biortech.2016.08.106. Epub 2016 Aug 31. Bioresour Technol. 2016. PMID: 27598569
-
Metabolic and gene expression changes triggered by nitrogen deprivation in the photoautotrophically grown microalgae Chlamydomonas reinhardtii and Coccomyxa sp. C-169.Phytochemistry. 2012 Mar;75:50-9. doi: 10.1016/j.phytochem.2011.12.007. Epub 2012 Jan 5. Phytochemistry. 2012. PMID: 22226037
-
Engineering fatty acid biosynthesis in microalgae for sustainable biodiesel.Curr Opin Chem Biol. 2013 Jun;17(3):496-505. doi: 10.1016/j.cbpa.2013.04.007. Epub 2013 May 14. Curr Opin Chem Biol. 2013. PMID: 23683348 Review.
-
Engineering photosynthetic organisms for the production of biohydrogen.Photosynth Res. 2015 Mar;123(3):241-53. doi: 10.1007/s11120-014-9991-x. Epub 2014 Mar 27. Photosynth Res. 2015. PMID: 24671643 Free PMC article. Review.
Cited by
-
Ectopic Expression of Perilla frutescens WRI1 Enhanced Storage Oil Accumulation in Nicotiana benthamiana Leaves.Plants (Basel). 2023 Feb 28;12(5):1081. doi: 10.3390/plants12051081. Plants (Basel). 2023. PMID: 36903941 Free PMC article.
-
Functional Characterization of Physcomitrella patens Glycerol-3-Phosphate Acyltransferase 9 and an Increase in Seed Oil Content in Arabidopsis by Its Ectopic Expression.Plants (Basel). 2019 Aug 13;8(8):284. doi: 10.3390/plants8080284. Plants (Basel). 2019. PMID: 31412690 Free PMC article.
-
Visualising the Emerging Platform of Using Microalgae as a Sustainable Bio-Factory for Healthy Lipid Production through Biocompatible AIE Probes.Biosensors (Basel). 2022 Mar 31;12(4):208. doi: 10.3390/bios12040208. Biosensors (Basel). 2022. PMID: 35448268 Free PMC article.
-
Draft genome sequence and detailed characterization of biofuel production by oleaginous microalga Scenedesmus quadricauda LWG002611.Biotechnol Biofuels. 2018 Nov 9;11:308. doi: 10.1186/s13068-018-1308-4. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 30455737 Free PMC article.
-
Characterization of a Haematococcus pluvialis Diacylglycerol Acyltransferase 1 and Its Potential in Unsaturated Fatty Acid-Rich Triacylglycerol Production.Front Plant Sci. 2021 Dec 7;12:771300. doi: 10.3389/fpls.2021.771300. eCollection 2021. Front Plant Sci. 2021. PMID: 34950166 Free PMC article.
References
-
- Adams, C. , Godfrey, V. , Wahlen, B. , Seefeldt, L. and Bugbee, B. (2013) Understanding precision nitrogen stress to optimize the growth and lipid content tradeoff in oleaginous green microalgae. Bioresour. Technol. 131, 188–194. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

