Integration of small RNAs, degradome and transcriptome sequencing in hyperaccumulator Sedum alfredii uncovers a complex regulatory network and provides insights into cadmium phytoremediation
- PMID: 26801211
- PMCID: PMC5066797
- DOI: 10.1111/pbi.12512
Integration of small RNAs, degradome and transcriptome sequencing in hyperaccumulator Sedum alfredii uncovers a complex regulatory network and provides insights into cadmium phytoremediation
Erratum in
-
Corrigendum.Plant Biotechnol J. 2021 Jan;19(1):206. doi: 10.1111/pbi.13518. Plant Biotechnol J. 2021. PMID: 33370514 Free PMC article. No abstract available.
Abstract
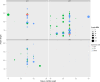
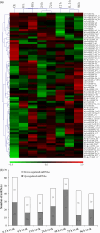
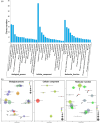
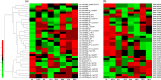
The hyperaccumulating ecotype of Sedum alfredii Hance is a cadmium (Cd)/zinc/lead co-hyperaccumulating species of Crassulaceae. It is a promising phytoremediation candidate accumulating substantial heavy metal ions without obvious signs of poisoning. However, few studies have focused on the regulatory roles of miRNAs and their targets in the hyperaccumulating ecotype of S. alfredii. Here, we combined analyses of the transcriptomics, sRNAs and the degradome to generate a comprehensive resource focused on identifying key regulatory miRNA-target circuits under Cd stress. A total of 87 721 unigenes and 356 miRNAs were identified by deep sequencing, and 79 miRNAs were differentially expressed under Cd stress. Furthermore, 754 target genes of 194 miRNAs were validated by degradome sequencing. A gene ontology (GO) enrichment analysis of differential miRNA targets revealed that auxin, redox-related secondary metabolism and metal transport pathways responded to Cd stress. An integrated analysis uncovered 39 pairs of miRNA targets that displayed negatively correlated expression profiles. Ten miRNA-target pairs also exhibited negative correlations according to a real-time quantitative PCR analysis. Moreover, a coexpression regulatory network was constructed based on profiles of differentially expressed genes. Two hub genes, ARF4 (auxin response factor 4) and AAP3 (amino acid permease 3), which might play central roles in the regulation of Cd-responsive genes, were uncovered. These results suggest that comprehensive analyses of the transcriptomics, sRNAs and the degradome provided a useful platform for investigating Cd hyperaccumulation in S. alfredii, and may provide new insights into the genetic engineering of phytoremediation.
Keywords: Sedum alfredii Hance; cadmium stress; coexpression network; integration analysis; phytoremediation.
© 2016 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures


Similar articles
-
Site-specific regulation of transcriptional responses to cadmium stress in the hyperaccumulator, Sedum alfredii: based on stem parenchymal and vascular cells.Plant Mol Biol. 2019 Mar;99(4-5):347-362. doi: 10.1007/s11103-019-00821-1. Epub 2019 Jan 14. Plant Mol Biol. 2019. PMID: 30644059
-
Transcriptomic analysis of cadmium stress response in the heavy metal hyperaccumulator Sedum alfredii Hance.PLoS One. 2013 Jun 3;8(6):e64643. doi: 10.1371/journal.pone.0064643. Print 2014. PLoS One. 2013. PMID: 23755133 Free PMC article.
-
Enhanced expression of SaHMA3 plays critical roles in Cd hyperaccumulation and hypertolerance in Cd hyperaccumulator Sedum alfredii Hance.Planta. 2016 Mar;243(3):577-89. doi: 10.1007/s00425-015-2429-7. Epub 2015 Nov 7. Planta. 2016. PMID: 26547194
-
Sedum alfredii Hance: A cadmium and zinc hyperaccumulating plant.Ecotoxicol Environ Saf. 2025 Jan 15;290:117588. doi: 10.1016/j.ecoenv.2024.117588. Epub 2024 Dec 24. Ecotoxicol Environ Saf. 2025. PMID: 39721422 Review.
-
Cadmium phytoextraction by Sedum alfredii and Sedum plumbizincicola: mechanisms, challenges and prospects.Int J Phytoremediation. 2025;27(6):852-860. doi: 10.1080/15226514.2025.2451714. Epub 2025 Jan 21. Int J Phytoremediation. 2025. PMID: 39838584 Review.
Cited by
-
Arsenic perception and signaling: The yet unexplored world.Front Plant Sci. 2022 Sep 2;13:993484. doi: 10.3389/fpls.2022.993484. eCollection 2022. Front Plant Sci. 2022. PMID: 36119603 Free PMC article. Review.
-
Identification and expression analysis of the GDSL esterase/lipase family genes, and the characterization of SaGLIP8 in Sedum alfredii Hance under cadmium stress.PeerJ. 2019 Apr 16;7:e6741. doi: 10.7717/peerj.6741. eCollection 2019. PeerJ. 2019. PMID: 31024765 Free PMC article.
-
Parallel analysis of RNA ends reveals global microRNA-mediated target RNA cleavage in maize.Plant J. 2022 Oct;112(1):268-283. doi: 10.1111/tpj.15943. Epub 2022 Aug 23. Plant J. 2022. PMID: 35962593 Free PMC article.
-
Endoplasmic reticulum stress-responsive microRNAs are involved in the regulation of abiotic stresses in wheat.Plant Cell Rep. 2023 Sep;42(9):1433-1452. doi: 10.1007/s00299-023-03040-7. Epub 2023 Jun 21. Plant Cell Rep. 2023. PMID: 37341828
-
Integration of small RNAs and transcriptome sequencing uncovers a complex regulatory network during vernalization and heading stages of orchardgrass (Dactylis glomerata L.).BMC Genomics. 2018 Oct 3;19(1):727. doi: 10.1186/s12864-018-5104-0. BMC Genomics. 2018. PMID: 30285619 Free PMC article.
References
-
- Arasimowicz‐Jelonek, M. , Floryszak‐Wieczorek, J. , Deckert, J. , Rucińska‐Sobkowiak, R. , Gzyl, J. , Pawlak‐Sprada, S. , Abramowski, D. et al. (2012) Nitric oxide implication in cadmium‐induced programmed cell death in roots and signaling response of yellow lupine plants. Plant Physiol. Biochem. 58, 124–134. - PubMed
-
- Boominathan, R. and Doran, P.M. (2003) Cadmium tolerance and antioxidative defenses in hairy roots of the cadmium hyperaccumulator, Thlaspi caerulescens . Biotechnol. Bioeng. 83, 158–167. - PubMed
-
- Brodersen, P. , Sakvarelidze‐Achard, L. , Bruun‐Rasmussen, M. , Dunoyer, P. , Yamamoto, Y.Y. , Sieburth, L. and Voinnet, O. (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science, 320, 1185–1190. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

