Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control
- PMID: 26801249
- PMCID: PMC4724118
- DOI: 10.1186/s12915-016-0229-6
Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control
Abstract
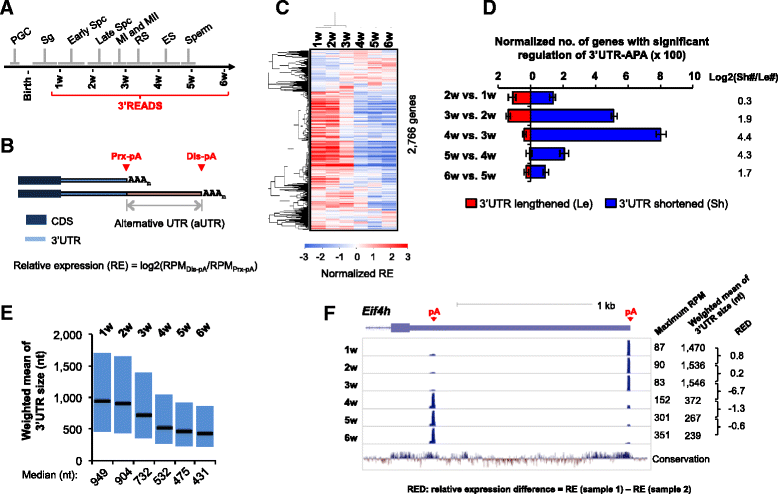
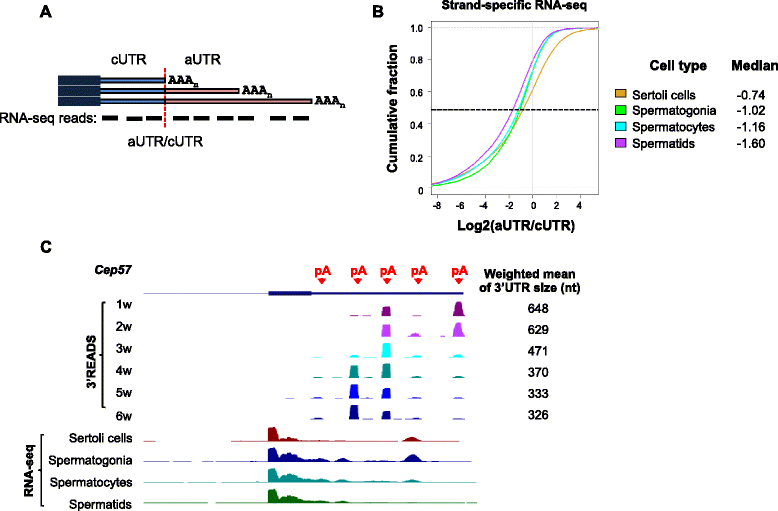
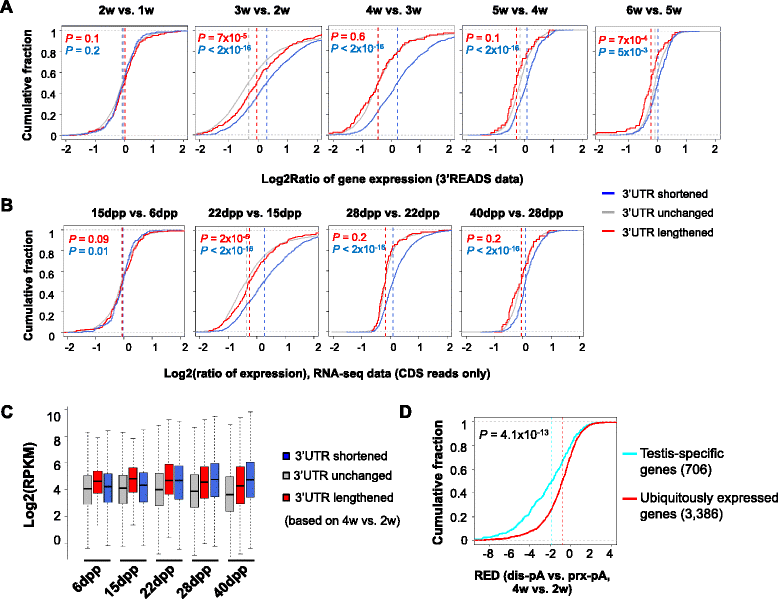
Background: Most mammalian genes display alternative cleavage and polyadenylation (APA). Previous studies have indicated preferential expression of APA isoforms with short 3' untranslated regions (3'UTRs) in testes.
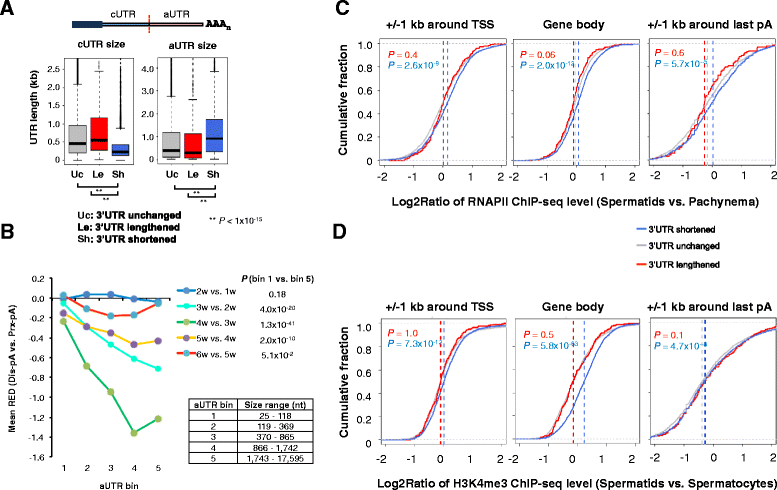
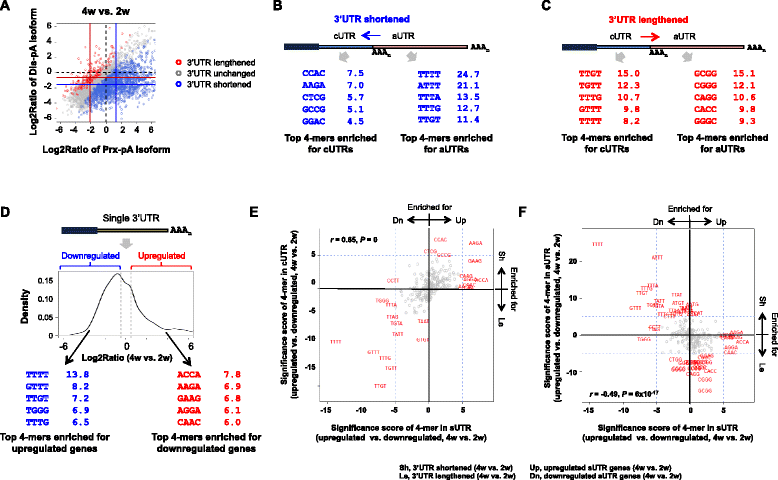
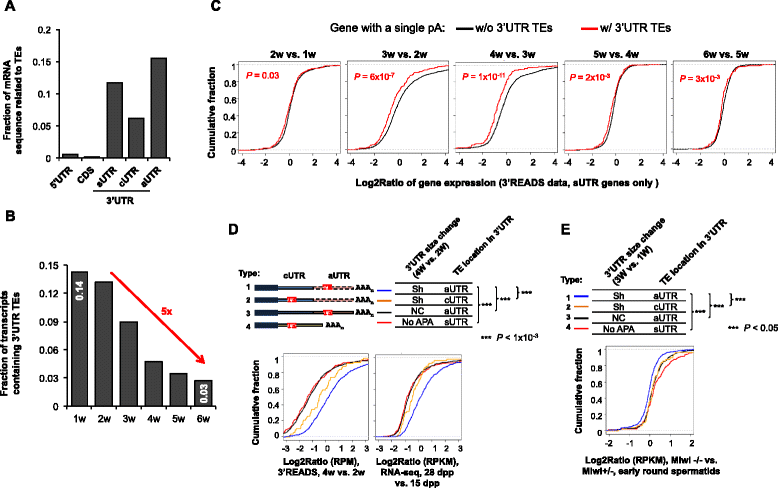
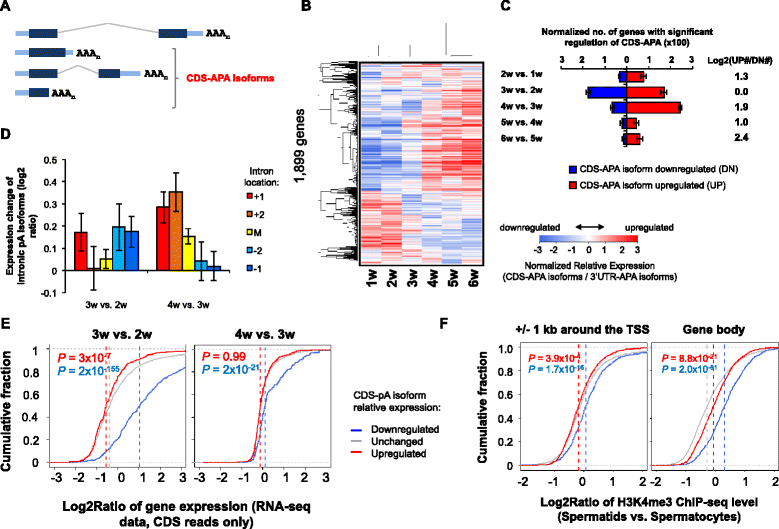
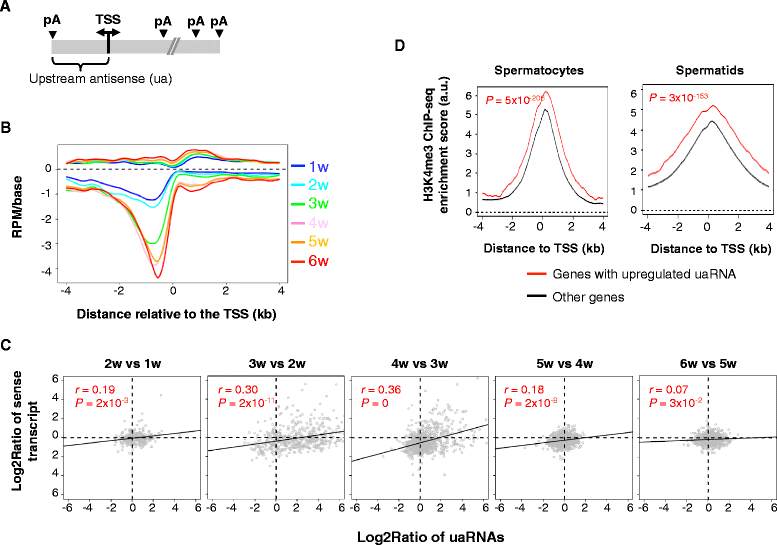
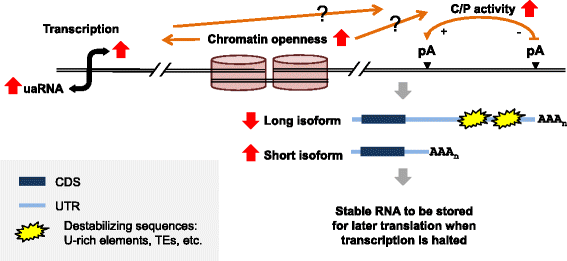
Results: By deep sequencing of the 3' end region of poly(A) + transcripts, we report widespread shortening of 3'UTR through APA during the first wave of spermatogenesis in mouse, with 3'UTR size being the shortest in spermatids. Using genes without APA as a control, we show that shortening of 3'UTR eliminates destabilizing elements, such as U-rich elements and transposable elements, which appear highly potent during spermatogenesis. We additionally found widespread regulation of APA events in introns and exons that can affect the coding sequence of transcripts and global activation of antisense transcripts upstream of the transcription start site, suggesting modulation of splicing and initiation of transcription during spermatogenesis. Importantly, genes that display significant 3'UTR shortening tend to have functions critical for further sperm maturation, and testis-specific genes display greater 3'UTR shortening than ubiquitously expressed ones, indicating functional relevance of APA to spermatogenesis. Interestingly, genes with shortened 3'UTRs tend to have higher RNA polymerase II and H3K4me3 levels in spermatids as compared to spermatocytes, features previously known to be associated with open chromatin state.
Conclusions: Our data suggest that open chromatin may create a favorable cis environment for 3' end processing, leading to global shortening of 3'UTR during spermatogenesis. mRNAs with shortened 3'UTRs are relatively stable thanks to evasion of powerful mRNA degradation mechanisms acting on 3'UTR elements. Stable mRNAs generated in spermatids may be important for protein production at later stages of sperm maturation, when transcription is globally halted.
Figures
Similar articles
-
Regulation and function of alternative polyadenylation in development and differentiation.RNA Biol. 2023 Jan;20(1):908-925. doi: 10.1080/15476286.2023.2275109. Epub 2023 Oct 31. RNA Biol. 2023. PMID: 37906624 Free PMC article. Review.
-
UPF2-Dependent Nonsense-Mediated mRNA Decay Pathway Is Essential for Spermatogenesis by Selectively Eliminating Longer 3'UTR Transcripts.PLoS Genet. 2016 May 5;12(5):e1005863. doi: 10.1371/journal.pgen.1005863. eCollection 2016 May. PLoS Genet. 2016. PMID: 27149259 Free PMC article.
-
Systematic identification of functional SNPs interrupting 3'UTR polyadenylation signals.PLoS Genet. 2020 Aug 17;16(8):e1008977. doi: 10.1371/journal.pgen.1008977. eCollection 2020 Aug. PLoS Genet. 2020. PMID: 32804959 Free PMC article.
-
Reprogramming of 3' untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types.PLoS One. 2009 Dec 23;4(12):e8419. doi: 10.1371/journal.pone.0008419. PLoS One. 2009. PMID: 20037631 Free PMC article.
-
The emerging theme of 3'UTR mRNA isoform regulation in reprogramming of cell metabolism.Biochem Soc Trans. 2023 Jun 28;51(3):1111-1119. doi: 10.1042/BST20221128. Biochem Soc Trans. 2023. PMID: 37171086 Free PMC article. Review.
Cited by
-
m6A-dependent biogenesis of circular RNAs in male germ cells.Cell Res. 2020 Mar;30(3):211-228. doi: 10.1038/s41422-020-0279-8. Epub 2020 Feb 11. Cell Res. 2020. PMID: 32047269 Free PMC article.
-
Quantification of transcript isoforms at the single-cell level using SCALPEL.Nat Commun. 2025 Jul 11;16(1):6402. doi: 10.1038/s41467-025-61118-0. Nat Commun. 2025. PMID: 40640129 Free PMC article.
-
Downregulation of CPSF6 leads to global mRNA 3' UTR shortening and enhanced antiviral immune responses.PLoS Pathog. 2024 Feb 28;20(2):e1012061. doi: 10.1371/journal.ppat.1012061. eCollection 2024 Feb. PLoS Pathog. 2024. PMID: 38416782 Free PMC article.
-
Macrophage differentiation is marked by increased abundance of the mRNA 3' end processing machinery, altered poly(A) site usage, and sensitivity to the level of CstF64.Front Immunol. 2023 Jan 25;14:1091403. doi: 10.3389/fimmu.2023.1091403. eCollection 2023. Front Immunol. 2023. PMID: 36761770 Free PMC article.
-
Regulation and function of alternative polyadenylation in development and differentiation.RNA Biol. 2023 Jan;20(1):908-925. doi: 10.1080/15476286.2023.2275109. Epub 2023 Oct 31. RNA Biol. 2023. PMID: 37906624 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

