Comparative genome-wide transcriptome analysis of Vitis vinifera responses to adapted and non-adapted strains of two-spotted spider mite, Tetranyhus urticae
- PMID: 26801623
- PMCID: PMC4724079
- DOI: 10.1186/s12864-016-2401-3
Comparative genome-wide transcriptome analysis of Vitis vinifera responses to adapted and non-adapted strains of two-spotted spider mite, Tetranyhus urticae
Abstract
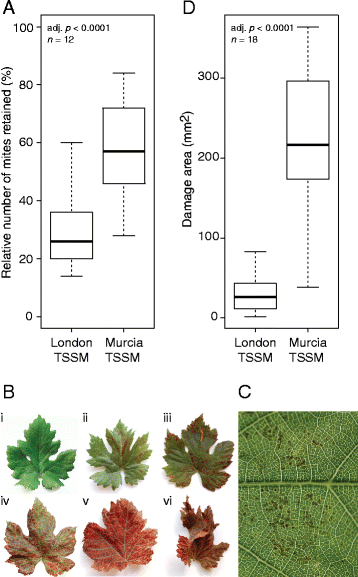
Background: The two-spotted spider mite, Tetranychus urticae, is an extreme generalist plant pest. Even though mites can feed on many plant species, local mite populations form host races that do not perform equally well on all potential hosts. An acquisition of the ability to evade plant defenses is fundamental for mite's ability to use a particular plant as a host. Thus, understanding the interactions between the plant and mites with different host adaptation status allows the identification of functional plant defenses and ways mites can evolve to avoid them.
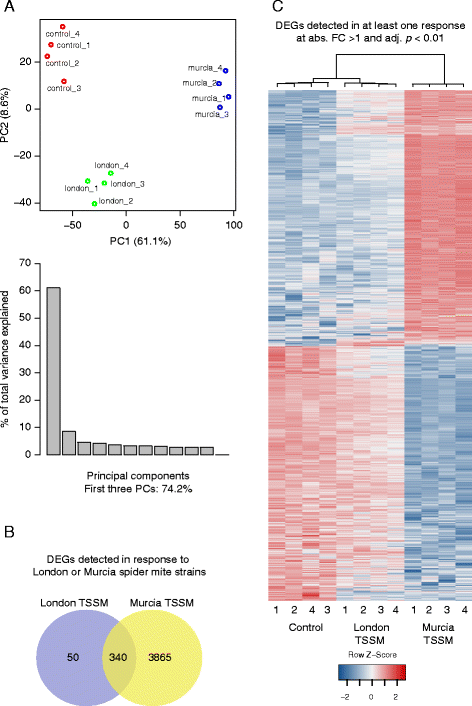
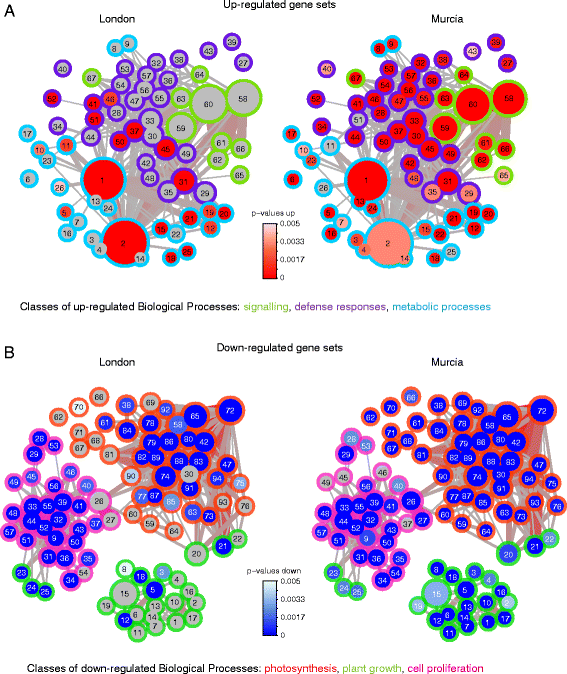
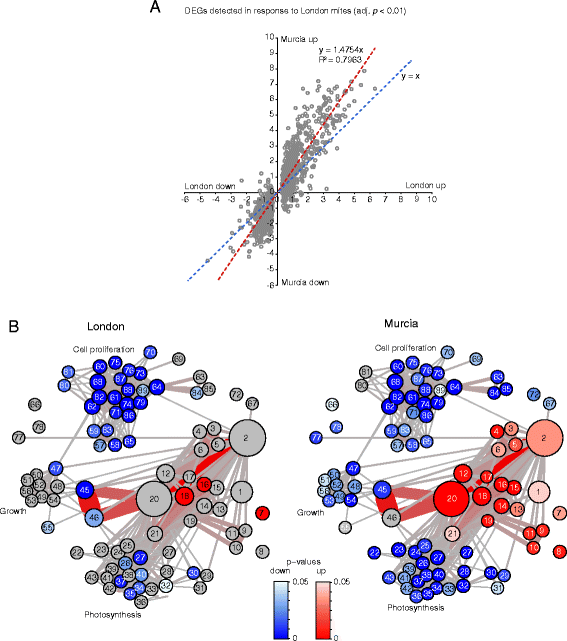
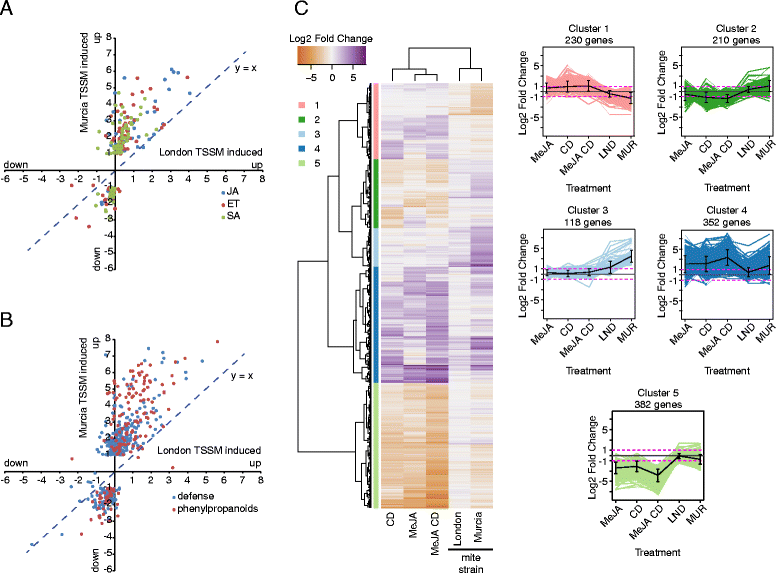
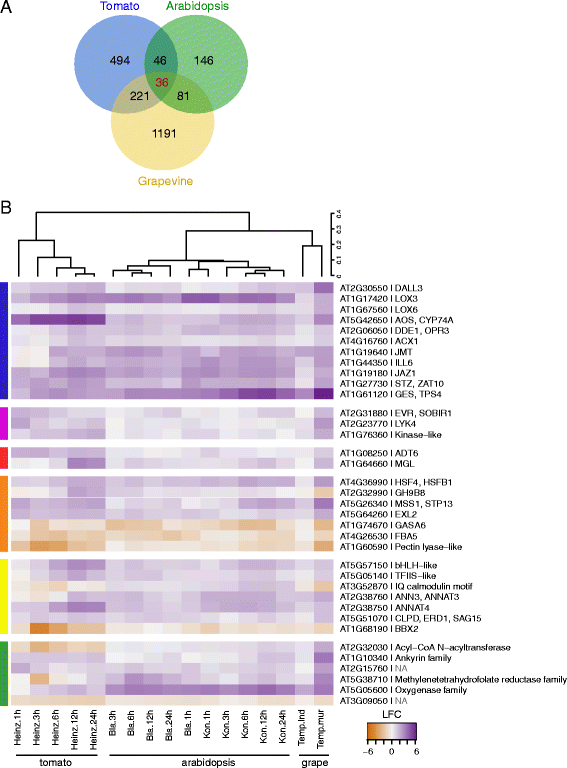
Results: The grapevine genome-wide transcriptional responses to spider mite strains that are non-adapted and adapted to grapevine as a host were examined. Comparative transcriptome analysis of grapevine responses to these mite strains identified the existence of weak responses induced by the feeding of the non-adapted strain. In contrast, strong but ineffective induced defenses were triggered upon feeding of the adapted strain. A comparative meta-analysis of Arabidopsis, tomato and grapevine responses to mite feeding identified a core of 36 highly conserved genes involved in the perception, regulation and metabolism that were commonly induced in all three species by mite herbivory.
Conclusions: This study describes the genome-wide grapevine transcriptional responses to herbivory of mite strains that differ in their ability to use grapevine as a host. It raises hypotheses whose testing will lead to our understanding of grapevine defenses and mite adaptations to them.
Figures
References
-
- Jeppson LR, Keifer HH, Baker EW. Mites injurious to economic plants. Berkeley: London: University of California Press; 1975.
-
- Spider Mites Web: a comprehensive database for the Tetranychidae. [http://www.montpellier.inra.fr/CBGP/spmweb]
-
- Martel C, Zhurov V, Navarro M, Martinez M, Cazaux M, Auger P, et al. Tomato Whole Genome Transcriptional Response to Tetranychus urticae Identifies Divergence of Spider Mite-Induced Responses Between Tomato and Arabidopsis. Mol Plant Microbe Interact. 2015;28(3):343–361. doi: 10.1094/MPMI-09-14-0291-FI. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

