Implementation of the Realized Genomic Relationship Matrix to Open-Pollinated White Spruce Family Testing for Disentangling Additive from Nonadditive Genetic Effects
- PMID: 26801647
- PMCID: PMC4777135
- DOI: 10.1534/g3.115.025957
Implementation of the Realized Genomic Relationship Matrix to Open-Pollinated White Spruce Family Testing for Disentangling Additive from Nonadditive Genetic Effects
Abstract
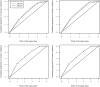
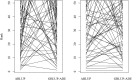
The open-pollinated (OP) family testing combines the simplest known progeny evaluation and quantitative genetics analyses as candidates' offspring are assumed to represent independent half-sib families. The accuracy of genetic parameter estimates is often questioned as the assumption of "half-sibling" in OP families may often be violated. We compared the pedigree- vs. marker-based genetic models by analysing 22-yr height and 30-yr wood density for 214 white spruce [Picea glauca (Moench) Voss] OP families represented by 1694 individuals growing on one site in Quebec, Canada. Assuming half-sibling, the pedigree-based model was limited to estimating the additive genetic variances which, in turn, were grossly overestimated as they were confounded by very minor dominance and major additive-by-additive epistatic genetic variances. In contrast, the implemented genomic pairwise realized relationship models allowed the disentanglement of additive from all nonadditive factors through genetic variance decomposition. The marker-based models produced more realistic narrow-sense heritability estimates and, for the first time, allowed estimating the dominance and epistatic genetic variances from OP testing. In addition, the genomic models showed better prediction accuracies compared to pedigree models and were able to predict individual breeding values for new individuals from untested families, which was not possible using the pedigree-based model. Clearly, the use of marker-based relationship approach is effective in estimating the quantitative genetic parameters of complex traits even under simple and shallow pedigree structure.
Keywords: GenPred; Mendelian sampling term; genetic variance decomposition; genomic selection; open-pollinated families; pedigree- and marker-based relationships; shared data resource.
Copyright © 2016 El-Dien et al.
Figures


Similar articles
-
Single-Step BLUP with Varying Genotyping Effort in Open-Pollinated Picea glauca.G3 (Bethesda). 2017 Mar 10;7(3):935-942. doi: 10.1534/g3.116.037895. G3 (Bethesda). 2017. PMID: 28122953 Free PMC article.
-
Multienvironment genomic variance decomposition analysis of open-pollinated Interior spruce (Picea glauca x engelmannii).Mol Breed. 2018;38(3):26. doi: 10.1007/s11032-018-0784-3. Epub 2018 Feb 15. Mol Breed. 2018. PMID: 29491726 Free PMC article.
-
Prediction accuracies for growth and wood attributes of interior spruce in space using genotyping-by-sequencing.BMC Genomics. 2015 May 9;16(1):370. doi: 10.1186/s12864-015-1597-y. BMC Genomics. 2015. PMID: 25956247 Free PMC article.
-
Genetic evaluation of dairy cattle using a simple heritable genetic ground.J Sci Food Agric. 2010 Aug 30;90(11):1765-73. doi: 10.1002/jsfa.4041. J Sci Food Agric. 2010. PMID: 20564310 Review.
-
Kinetics genetics: Incorporating the concept of genomic balance into an understanding of quantitative traits.Plant Sci. 2016 Apr;245:128-34. doi: 10.1016/j.plantsci.2016.02.002. Epub 2016 Feb 6. Plant Sci. 2016. PMID: 26940497 Review.
Cited by
-
Genomic evaluation for breeding and genetic management in Cordia africana, a multipurpose tropical tree species.BMC Genomics. 2024 Jan 2;25(1):9. doi: 10.1186/s12864-023-09907-z. BMC Genomics. 2024. PMID: 38166623 Free PMC article.
-
Effect of Hidden Relatedness on Single-Step Genetic Evaluation in an Advanced Open-Pollinated Breeding Program.J Hered. 2018 Oct 31;109(7):802-810. doi: 10.1093/jhered/esy051. J Hered. 2018. PMID: 30285150 Free PMC article.
-
Genomic dissection of additive and non-additive genetic effects and genomic prediction in an open-pollinated family test of Japanese larch.BMC Genomics. 2024 Jan 2;25(1):11. doi: 10.1186/s12864-023-09891-4. BMC Genomics. 2024. PMID: 38166605 Free PMC article.
-
Effect of number of annual rings and tree ages on genomic predictive ability for solid wood properties of Norway spruce.BMC Genomics. 2020 Apr 25;21(1):323. doi: 10.1186/s12864-020-6737-3. BMC Genomics. 2020. PMID: 32334511 Free PMC article.
-
Single-Step BLUP with Varying Genotyping Effort in Open-Pollinated Picea glauca.G3 (Bethesda). 2017 Mar 10;7(3):935-942. doi: 10.1534/g3.116.037895. G3 (Bethesda). 2017. PMID: 28122953 Free PMC article.
References
-
- Askew G. R., El-Kassaby Y. A., 1994. Estimation of relationship coefficients among progeny derived from wind-pollinated orchard seeds. Theor. Appl. Genet. 88: 267–272. - PubMed
-
- Avendano S., Woolliams J. A., Villanueva B., 2004. Mendelian sampling terms as a selective advantage in optimum breeding schemes with restrictions on the rate of inbreeding. Genet. Res. 83: 55–64. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
