Metabolic stress is a barrier to Epstein-Barr virus-mediated B-cell immortalization
- PMID: 26802124
- PMCID: PMC4760815
- DOI: 10.1073/pnas.1517141113
Metabolic stress is a barrier to Epstein-Barr virus-mediated B-cell immortalization
Abstract
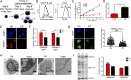
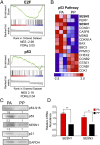
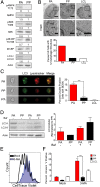
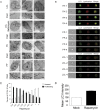
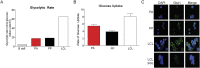
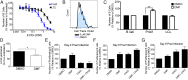
Epstein-Barr virus (EBV) is an oncogenic herpesvirus that has been causally linked to the development of B-cell and epithelial malignancies. Early after infection, EBV induces a transient period of hyperproliferation that is suppressed by the activation of the DNA damage response and a G1/S-phase growth arrest. This growth arrest prevents long-term outgrowth of the majority of infected cells. We developed a method to isolate and characterize infected cells that arrest after this early burst of proliferation and integrated gene expression and metabolic profiling to gain a better understanding of the pathways that attenuate immortalization. We found that the arrested cells have a reduced level of mitochondrial respiration and a decrease in the expression of genes involved in the TCA cycle and oxidative phosphorylation. Indeed, the growth arrest in early infected cells could be rescued by supplementing the TCA cycle. Arrested cells were characterized by an increase in the expression of p53 pathway gene targets, including sestrins leading to activation of AMPK, a reduction in mTOR signaling, and, consequently, elevated autophagy that was important for cell survival. Autophagy was also critical to maintain early hyperproliferation during metabolic stress. Finally, in assessing the metabolic changes from early infection to long-term outgrowth, we found concomitant increases in glucose import and surface glucose transporter 1 (GLUT1) levels, leading to elevated glycolysis, oxidative phosphorylation, and suppression of basal autophagy. Our study demonstrates that oncogene-induced senescence triggered by a combination of metabolic and genotoxic stress acts as an intrinsic barrier to EBV-mediated transformation.
Keywords: B cell; Epstein–Barr virus; autophagy; metabolism; oncogene-induced senescence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Latent membrane protein 1 of Epstein-Barr virus plays an important role in the serum starvation resistance of Epstein-Barr virus-immortalized B lymphocytes.J Cell Biochem. 2004 Mar 1;91(4):777-85. doi: 10.1002/jcb.10776. J Cell Biochem. 2004. PMID: 14991769
-
An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells.Cell Host Microbe. 2010 Dec 16;8(6):510-22. doi: 10.1016/j.chom.2010.11.004. Cell Host Microbe. 2010. PMID: 21147465 Free PMC article.
-
The Epstein-Barr virus nuclear antigen-1 upregulates the cellular antioxidant defense to enable B-cell growth transformation and immortalization.Oncogene. 2020 Jan;39(3):603-616. doi: 10.1038/s41388-019-1003-3. Epub 2019 Sep 11. Oncogene. 2020. PMID: 31511648 Free PMC article.
-
Regulation of Telomere Homeostasis during Epstein-Barr virus Infection and Immortalization.Viruses. 2017 Aug 9;9(8):217. doi: 10.3390/v9080217. Viruses. 2017. PMID: 28792435 Free PMC article. Review.
-
Aberrant mTOR activation in senescence and aging: A mitochondrial stress response?Exp Gerontol. 2015 Aug;68:66-70. doi: 10.1016/j.exger.2014.11.004. Epub 2014 Nov 6. Exp Gerontol. 2015. PMID: 25449851 Free PMC article. Review.
Cited by
-
SARS-CoV-2 Causes Lung Inflammation through Metabolic Reprogramming and RAGE.Viruses. 2022 May 6;14(5):983. doi: 10.3390/v14050983. Viruses. 2022. PMID: 35632725 Free PMC article.
-
Contribution of viral and bacterial infections to senescence and immunosenescence.Front Cell Infect Microbiol. 2023 Sep 11;13:1229098. doi: 10.3389/fcimb.2023.1229098. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37753486 Free PMC article. Review.
-
Direct Evidence of Abortive Lytic Infection-Mediated Establishment of Epstein-Barr Virus Latency During B-Cell Infection.Front Microbiol. 2021 Jan 21;11:575255. doi: 10.3389/fmicb.2020.575255. eCollection 2020. Front Microbiol. 2021. PMID: 33613459 Free PMC article.
-
Epstein-Barr-Virus-Driven Cardiolipin Synthesis Sustains Metabolic Remodeling During B-cell Lymphomagenesis.Res Sq [Preprint]. 2024 Apr 8:rs.3.rs-4013392. doi: 10.21203/rs.3.rs-4013392/v1. Res Sq. 2024. Update in: Sci Adv. 2025 Jan 31;11(5):eadr8837. doi: 10.1126/sciadv.adr8837. PMID: 38659762 Free PMC article. Updated. Preprint.
-
EBV and Apoptosis: The Viral Master Regulator of Cell Fate?Viruses. 2017 Nov 13;9(11):339. doi: 10.3390/v9110339. Viruses. 2017. PMID: 29137176 Free PMC article. Review.
References
-
- Rickinson A, Kieff E. In: Epstein-Barr Virus Fields Virology. 5th Ed. Knipe DM, Howley PM, editors. Lippincott, Williams, and Wilkins; Philadelphia: 2007. pp. 2603–2654.
-
- Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181(2):595–608. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

