Cross-Reactive and Potent Neutralizing Antibody Responses in Human Survivors of Natural Ebolavirus Infection
- PMID: 26806128
- PMCID: PMC4733404
- DOI: 10.1016/j.cell.2015.12.022
Cross-Reactive and Potent Neutralizing Antibody Responses in Human Survivors of Natural Ebolavirus Infection
Abstract
Recent studies have suggested that antibody-mediated protection against the Ebolaviruses may be achievable, but little is known about whether or not antibodies can confer cross-reactive protection against viruses belonging to diverse Ebolavirus species, such as Ebola virus (EBOV), Sudan virus (SUDV), and Bundibugyo virus (BDBV). We isolated a large panel of human monoclonal antibodies (mAbs) against BDBV glycoprotein (GP) using peripheral blood B cells from survivors of the 2007 BDBV outbreak in Uganda. We determined that a large proportion of mAbs with potent neutralizing activity against BDBV bind to the glycan cap and recognize diverse epitopes within this major antigenic site. We identified several glycan cap-specific mAbs that neutralized multiple ebolaviruses, including SUDV, and a cross-reactive mAb that completely protected guinea pigs from the lethal challenge with heterologous EBOV. Our results provide a roadmap to develop a single antibody-based treatment effective against multiple Ebolavirus infections.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
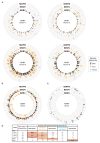

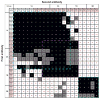


References
-
- Bray M, Davis K, Geisbert T, Schmaljohn C, Huggins J. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J Infect Dis. 1998;178:651–661. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1 TR000445/TR/NCATS NIH HHS/United States
- HHSN272201400058C/PHS HHS/United States
- P30 CA68485/CA/NCI NIH HHS/United States
- U19 AI109711/AI/NIAID NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R01 AI067927/AI/NIAID NIH HHS/United States
- U19 AI109762/AI/NIAID NIH HHS/United States
- 2 UL1 TR000445-06/TR/NCATS NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- HHSN272201400058C/AI/NIAID NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- DK058404/DK/NIDDK NIH HHS/United States
- UL1 RR024975-01/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

