Within-host evolution of bacterial pathogens
- PMID: 26806595
- PMCID: PMC5053366
- DOI: 10.1038/nrmicro.2015.13
Within-host evolution of bacterial pathogens
Abstract
Whole-genome sequencing has opened the way for investigating the dynamics and genomic evolution of bacterial pathogens during the colonization and infection of humans. The application of this technology to the longitudinal study of adaptation in an infected host--in particular, the evolution of drug resistance and host adaptation in patients who are chronically infected with opportunistic pathogens--has revealed remarkable patterns of convergent evolution, suggestive of an inherent repeatability of evolution. In this Review, we describe how these studies have advanced our understanding of the mechanisms and principles of within-host genome evolution, and we consider the consequences of findings such as a potent adaptive potential for pathogenicity. Finally, we discuss the possibility that genomics may be used in the future to predict the clinical progression of bacterial infections and to suggest the best option for treatment.
Figures
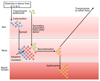
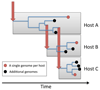

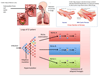
References
-
-
Lieberman et al (2011) Nat Genet
- [Study of a B. dolosa outbreak in cystic fibrosis patients revealed evidence for adaptation to the host in the form of convergent evolution across multiple patients at genes encoding antibiotic resistance and bacterial membrane composition.]
-
-
-
Markussen et al (2014) mBio
- [Investigation of a 32-year persistent P. aeruginosa DK1 infection showed diversification and co-existence of sub-lineages with distinct functional and genomic signatures and rates of evolution that may occupy different niches within the cystic fibrosis airway.]
-
-
-
Marvig et al (2013) PLoS Genet
- [Evolutionary analysis of the P. aeruginosa DK2 lineage over 38 years identified pathoadaptive mutations occurring independently in multiple patients in genes relating to antibiotic resistance, cell envelope and regulatory functions.]
-
-
-
Elholm et al (2014) Genome Biol
- [First documented case of extensively drug resistant (XDR) M. tuberculosis strain evolved from a susceptible ancestor within a single patient. Resistance for most drugs evolved multiple times, with ultimately one lineage prevailing.]
-
-
-
Kennemann et al (2011) PNAS
- [Examination of five longitudinally sampled patients infected with H. pylori, detailing extensive mutation and recombination within individual hosts.]
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

