HIV-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases
- PMID: 26808478
- PMCID: PMC4836387
- DOI: 10.1021/acschembio.5b00948
HIV-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases
Abstract
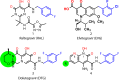
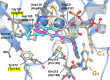
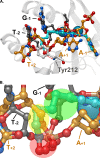
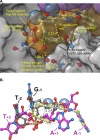
HIV integrase (IN) strand transfer inhibitors (INSTIs) are among the newest anti-AIDS drugs; however, mutant forms of IN can confer resistance. We developed noncytotoxic naphthyridine-containing INSTIs that retain low nanomolar IC50 values against HIV-1 variants harboring all of the major INSTI-resistant mutations. We found by analyzing crystal structures of inhibitors bound to the IN from the prototype foamy virus (PFV) that the most successful inhibitors show striking mimicry of the bound viral DNA prior to 3'-processing and the bound host DNA prior to strand transfer. Using this concept of "bi-substrate mimicry," we developed a new broadly effective inhibitor that not only mimics aspects of both the bound target and viral DNA but also more completely fills the space they would normally occupy. Maximizing shape complementarity and recapitulating structural components encompassing both of the IN DNA substrates could serve as a guiding principle for the development of new INSTIs.
Conflict of interest statement
The authors declare the following competing financial interest(s): Compounds described in the paper are included in pending or filed patent applications.
Figures
Similar articles
-
Close-up: HIV/SIV intasome structures shed new light on integrase inhibitor binding and viral escape mechanisms.FEBS J. 2021 Jan;288(2):427-433. doi: 10.1111/febs.15438. Epub 2020 Jun 22. FEBS J. 2021. PMID: 32506843 Free PMC article. Review.
-
Selectivity for strand-transfer over 3'-processing and susceptibility to clinical resistance of HIV-1 integrase inhibitors are driven by key enzyme-DNA interactions in the active site.Nucleic Acids Res. 2016 Aug 19;44(14):6896-906. doi: 10.1093/nar/gkw592. Epub 2016 Jul 1. Nucleic Acids Res. 2016. PMID: 27369381 Free PMC article.
-
Structure-Guided Optimization of HIV Integrase Strand Transfer Inhibitors.J Med Chem. 2017 Sep 14;60(17):7315-7332. doi: 10.1021/acs.jmedchem.7b00596. Epub 2017 Aug 10. J Med Chem. 2017. PMID: 28737946 Free PMC article.
-
Altered viral fitness and drug susceptibility in HIV-1 carrying mutations that confer resistance to nonnucleoside reverse transcriptase and integrase strand transfer inhibitors.J Virol. 2014 Aug;88(16):9268-76. doi: 10.1128/JVI.00695-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899199 Free PMC article.
-
Different Pathways Conferring Integrase Strand-Transfer Inhibitors Resistance.Viruses. 2022 Nov 22;14(12):2591. doi: 10.3390/v14122591. Viruses. 2022. PMID: 36560595 Free PMC article. Review.
Cited by
-
Probing Resistance Mutations in Retroviral Integrases by Direct Measurement of Dolutegravir Fluorescence.Sci Rep. 2017 Oct 25;7(1):14067. doi: 10.1038/s41598-017-14564-w. Sci Rep. 2017. PMID: 29070877 Free PMC article.
-
Close-up: HIV/SIV intasome structures shed new light on integrase inhibitor binding and viral escape mechanisms.FEBS J. 2021 Jan;288(2):427-433. doi: 10.1111/febs.15438. Epub 2020 Jun 22. FEBS J. 2021. PMID: 32506843 Free PMC article. Review.
-
Enhanced Solubility and Bioavailability of Dolutegravir by Solid Dispersion Method: In Vitro and In Vivo Evaluation-a Potential Approach for HIV Therapy.AAPS PharmSciTech. 2021 Apr 9;22(3):127. doi: 10.1208/s12249-021-01995-y. AAPS PharmSciTech. 2021. PMID: 33835317
-
Retroviral integrase: Structure, mechanism, and inhibition.Enzymes. 2021;50:249-300. doi: 10.1016/bs.enz.2021.06.007. Epub 2021 Aug 23. Enzymes. 2021. PMID: 34861940 Free PMC article.
-
Comparative Analyses of Antiviral Potencies of Second-Generation Integrase Strand Transfer Inhibitors (INSTIs) and the Developmental Compound 4d Against a Panel of Integrase Quadruple Mutants.Viruses. 2025 Jan 16;17(1):121. doi: 10.3390/v17010121. Viruses. 2025. PMID: 39861910 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

