Frequent chloroplast RNA editing in early-branching flowering plants: pilot studies on angiosperm-wide coexistence of editing sites and their nuclear specificity factors
- PMID: 26809609
- PMCID: PMC4727281
- DOI: 10.1186/s12862-016-0589-0
Frequent chloroplast RNA editing in early-branching flowering plants: pilot studies on angiosperm-wide coexistence of editing sites and their nuclear specificity factors
Abstract
Background: RNA editing by cytidine-to-uridine conversions is an essential step of RNA maturation in plant organelles. Some 30-50 sites of C-to-U RNA editing exist in chloroplasts of flowering plant models like Arabidopsis, rice or tobacco. We now predicted significantly more RNA editing in chloroplasts of early-branching angiosperm genera like Amborella, Calycanthus, Ceratophyllum, Chloranthus, Illicium, Liriodendron, Magnolia, Nuphar and Zingiber. Nuclear-encoded RNA-binding pentatricopeptide repeat (PPR) proteins are key editing factors expected to coevolve with their cognate RNA editing sites in the organelles.
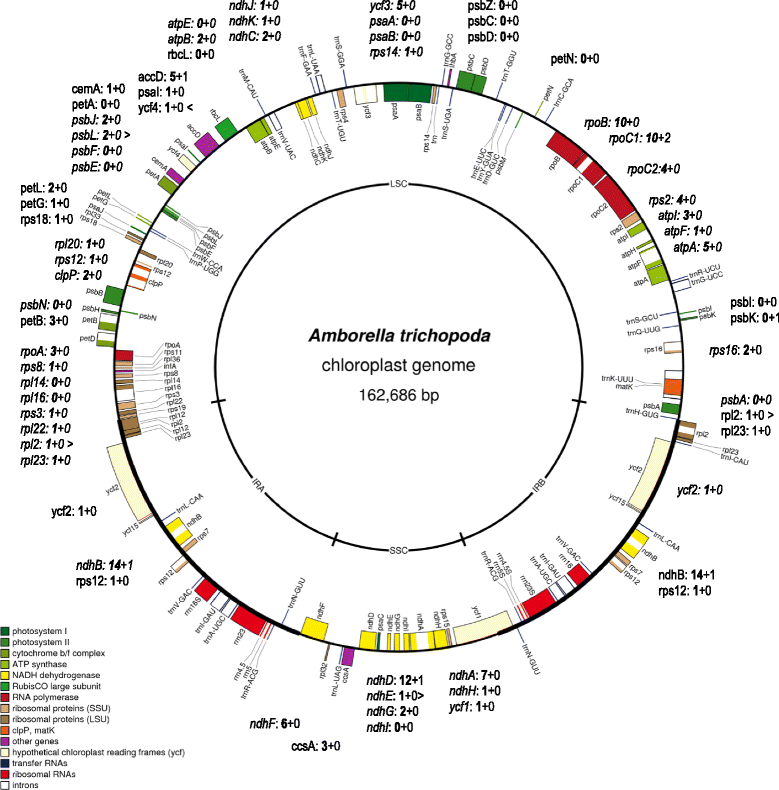
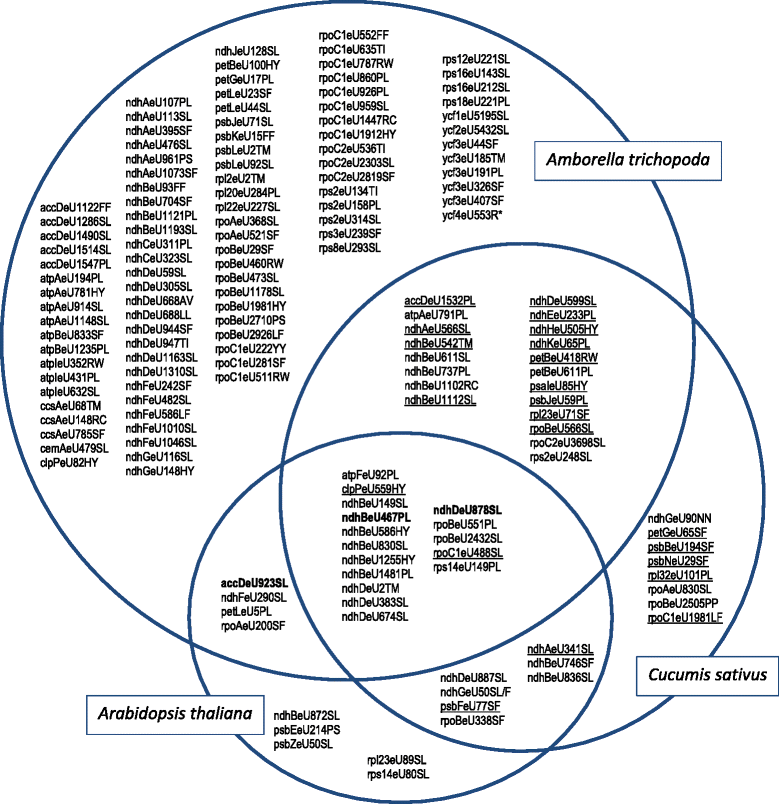
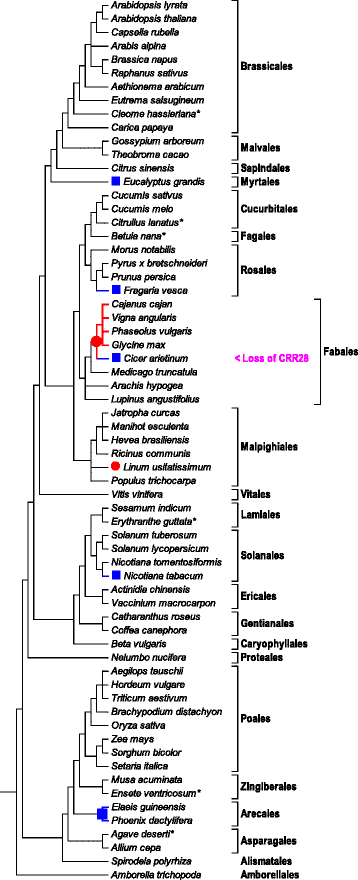
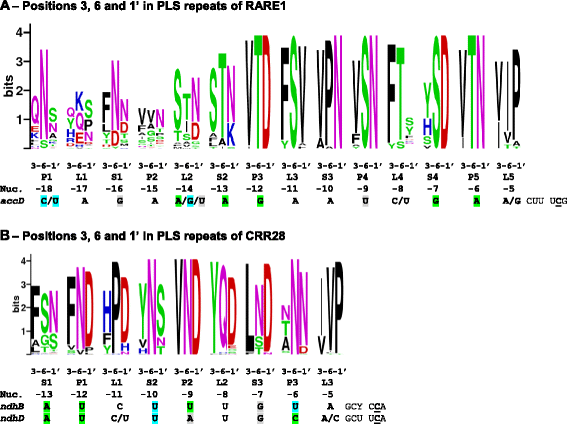
Results: With an extensive chloroplast transcriptome study we identified 138 sites of RNA editing in Amborella trichopoda, approximately the 3- to 4-fold of cp editing in Arabidopsis thaliana or Oryza sativa. Selected cDNA studies in the other early-branching flowering plant taxa furthermore reveal a high diversity of early angiosperm RNA editomes. Many of the now identified editing sites in Amborella have orthologues in ferns, lycophytes or hornworts. We investigated the evolution of CRR28 and RARE1, two known Arabidopsis RNA editing factors responsible for cp editing events ndhBeU467PL, ndhDeU878SL and accDeU794SL, respectively, all of which we now found conserved in Amborella. In a phylogenetically wide sampling of 65 angiosperm genomes we find evidence for only one single loss of CRR28 in chickpea but several independent losses of RARE1, perfectly congruent with the presence of their cognate editing sites in the respective cpDNAs.
Conclusion: Chloroplast RNA editing is much more abundant in early-branching than in widely investigated model flowering plants. RNA editing specificity factors can be traced back for more than 120 million years of angiosperm evolution and show highly divergent patterns of evolutionary losses, matching the presence of their target editing events.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

