Impact of germline and somatic missense variations on drug binding sites
- PMID: 26810135
- PMCID: PMC5380835
- DOI: 10.1038/tpj.2015.97
Impact of germline and somatic missense variations on drug binding sites
Abstract
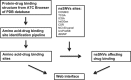
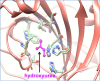
Advancements in next-generation sequencing (NGS) technologies are generating a vast amount of data. This exacerbates the current challenge of translating NGS data into actionable clinical interpretations. We have comprehensively combined germline and somatic nonsynonymous single-nucleotide variations (nsSNVs) that affect drug binding sites in order to investigate their prevalence. The integrated data thus generated in conjunction with exome or whole-genome sequencing can be used to identify patients who may not respond to a specific drug because of alterations in drug binding efficacy due to nsSNVs in the target protein's gene. To identify the nsSNVs that may affect drug binding, protein-drug complex structures were retrieved from Protein Data Bank (PDB) followed by identification of amino acids in the protein-drug binding sites using an occluded surface method. Then, the germline and somatic mutations were mapped to these amino acids to identify which of these alter protein-drug binding sites. Using this method we identified 12 993 amino acid-drug binding sites across 253 unique proteins bound to 235 unique drugs. The integration of amino acid-drug binding sites data with both germline and somatic nsSNVs data sets revealed 3133 nsSNVs affecting amino acid-drug binding sites. In addition, a comprehensive drug target discovery was conducted based on protein structure similarity and conservation of amino acid-drug binding sites. Using this method, 81 paralogs were identified that could serve as alternative drug targets. In addition, non-human mammalian proteins bound to drugs were used to identify 142 homologs in humans that can potentially bind to drugs. In the current protein-drug pairs that contain somatic mutations within their binding site, we identified 85 proteins with significant differential gene expression changes associated with specific cancer types. Information on protein-drug binding predicted drug target proteins and prevalence of both somatic and germline nsSNVs that disrupt these binding sites can provide valuable knowledge for personalized medicine treatment. A web portal is available where nsSNVs from individual patient can be checked by scanning against DrugVar to determine whether any of the SNVs affect the binding of any drug in the database.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Impact of Nonsynonymous Single-Nucleotide Variations on Post-Translational Modification Sites in Human Proteins.Methods Mol Biol. 2017;1558:159-190. doi: 10.1007/978-1-4939-6783-4_8. Methods Mol Biol. 2017. PMID: 28150238
-
A method to reduce ancestry related germline false positives in tumor only somatic variant calling.BMC Med Genomics. 2017 Oct 19;10(1):61. doi: 10.1186/s12920-017-0296-8. BMC Med Genomics. 2017. PMID: 29052513 Free PMC article.
-
Personalized genomic analyses for cancer mutation discovery and interpretation.Sci Transl Med. 2015 Apr 15;7(283):283ra53. doi: 10.1126/scitranslmed.aaa7161. Sci Transl Med. 2015. PMID: 25877891 Free PMC article.
-
Progressing the utilisation of pharmacogenetics and pharmacogenomics into clinical care.Pathology. 2013 Jun;45(4):357-70. doi: 10.1097/PAT.0b013e328360b66e. Pathology. 2013. PMID: 23594690 Review.
-
Prediction of drug response and adverse drug reactions: From twin studies to Next Generation Sequencing.Eur J Pharm Sci. 2019 Mar 15;130:65-77. doi: 10.1016/j.ejps.2019.01.024. Epub 2019 Jan 23. Eur J Pharm Sci. 2019. PMID: 30684656 Review.
Cited by
-
Rare genetic variability in human drug target genes modulates drug response and can guide precision medicine.Sci Adv. 2021 Sep 3;7(36):eabi6856. doi: 10.1126/sciadv.abi6856. Epub 2021 Sep 1. Sci Adv. 2021. PMID: 34516913 Free PMC article.
-
Pharmacogenomics-Based Detection of Variants Involved in Pain, Anti-inflammatory and Immunomodulating Agents Pathways by Whole Exome Sequencing and Deep in Silico Investigations Revealed Novel Chemical Carcinogenesis and Cancer Risks.Iran J Med Sci. 2025 Feb 1;50(2):98-111. doi: 10.30476/ijms.2024.101852.3450. eCollection 2025 Feb. Iran J Med Sci. 2025. PMID: 40026294 Free PMC article.
-
Expanded analysis of secondary germline findings from matched tumor/normal sequencing identifies additional clinically significant mutations.JCO Precis Oncol. 2019;3:PO.18.00143. doi: 10.1200/PO.18.00143. Epub 2019 Apr 11. JCO Precis Oncol. 2019. PMID: 31517177 Free PMC article.
-
In silico analysis of PFN1 related to amyotrophic lateral sclerosis.PLoS One. 2019 Jun 19;14(6):e0215723. doi: 10.1371/journal.pone.0215723. eCollection 2019. PLoS One. 2019. PMID: 31216283 Free PMC article.
References
-
- Venter JC, Levy S, Stockwell T, Remington K, Halpern A. Massive parallelism, randomness and genomic advances. Nat Genet 2003; 33: 219–227. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

