RNA-seq analysis of the gonadal transcriptome during Alligator mississippiensis temperature-dependent sex determination and differentiation
- PMID: 26810479
- PMCID: PMC4727388
- DOI: 10.1186/s12864-016-2396-9
RNA-seq analysis of the gonadal transcriptome during Alligator mississippiensis temperature-dependent sex determination and differentiation
Abstract
Background: The American alligator (Alligator mississippiensis) displays temperature-dependent sex determination (TSD), in which incubation temperature during embryonic development determines the sexual fate of the individual. However, the molecular mechanisms governing this process remain a mystery, including the influence of initial environmental temperature on the comprehensive gonadal gene expression patterns occurring during TSD.
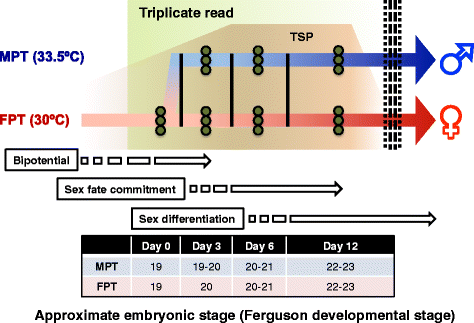
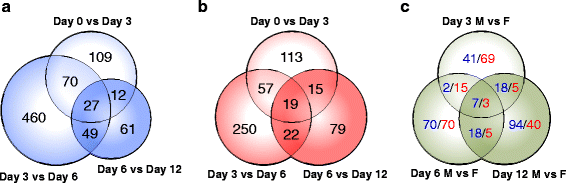
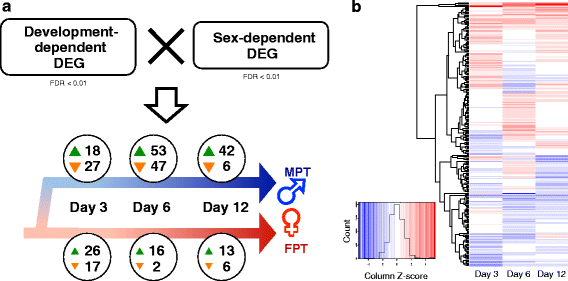
Results: Our characterization of transcriptomes during alligator TSD allowed us to identify novel candidate genes involved in TSD initiation. High-throughput RNA sequencing (RNA-seq) was performed on gonads collected from A. mississippiensis embryos incubated at both a male and a female producing temperature (33.5 °C and 30 °C, respectively) in a time series during sexual development. RNA-seq yielded 375.2 million paired-end reads, which were mapped and assembled, and used to characterize differential gene expression. Changes in the transcriptome occurring as a function of both development and sexual differentiation were extensively profiled. Forty-one differentially expressed genes were detected in response to incubation at male producing temperature, and included genes such as Wnt signaling factor WNT11, histone demethylase KDM6B, and transcription factor C/EBPA. Furthermore, comparative analysis of development- and sex-dependent differential gene expression revealed 230 candidate genes involved in alligator sex determination and differentiation, and early details of the suspected male-fate commitment were profiled. We also discovered sexually dimorphic expression of uncharacterized ncRNAs and other novel elements, such as unique expression patterns of HEMGN and ARX. Twenty-five of the differentially expressed genes identified in our analysis were putative transcriptional regulators, among which were MYBL2, MYCL, and HOXC10, in addition to conventional sex differentiation genes such as SOX9, and FOXL2. Inferred gene regulatory network was constructed, and the gene-gene and temperature-gene interactions were predicted.
Conclusions: Gonadal global gene expression kinetics during sex determination has been extensively profiled for the first time in a TSD species. These findings provide insights into the genetic framework underlying TSD, and expand our current understanding of the developmental fate pathways during vertebrate sex determination.
Figures


Similar articles
-
Molecular cloning of anti-Müllerian hormone from the American alligator, Alligator mississippiensis.Mol Cell Endocrinol. 2011 Feb 20;333(2):190-9. doi: 10.1016/j.mce.2010.12.025. Epub 2010 Dec 25. Mol Cell Endocrinol. 2011. PMID: 21187121
-
Differential incubation temperatures result in dimorphic DNA methylation patterning of the SOX9 and aromatase promoters in gonads of alligator (Alligator mississippiensis) embryos.Biol Reprod. 2014 Jan 9;90(1):2. doi: 10.1095/biolreprod.113.111468. Print 2014 Jan. Biol Reprod. 2014. PMID: 24227754
-
The influence of thermal signals during embryonic development on intrasexual and sexually dimorphic gene expression and circulating steroid hormones in American alligator hatchlings (Alligator mississippiensis).Gen Comp Endocrinol. 2016 Nov 1;238:47-54. doi: 10.1016/j.ygcen.2016.04.011. Epub 2016 Apr 11. Gen Comp Endocrinol. 2016. PMID: 27080549
-
Environmental sex determination mechanisms in reptiles.Sex Dev. 2013;7(1-3):95-103. doi: 10.1159/000341936. Epub 2012 Aug 31. Sex Dev. 2013. PMID: 22948613 Review.
-
Yolk steroid hormones and sex determination in reptiles with TSD.Gen Comp Endocrinol. 2003 Jul;132(3):349-55. doi: 10.1016/s0016-6480(03)00098-4. Gen Comp Endocrinol. 2003. PMID: 12849957 Review.
Cited by
-
Identification of General Patterns of Sex-Biased Expression in Daphnia, a Genus with Environmental Sex Determination.G3 (Bethesda). 2018 May 4;8(5):1523-1533. doi: 10.1534/g3.118.200174. G3 (Bethesda). 2018. PMID: 29535148 Free PMC article.
-
Differential Mitochondrial Genome Expression of Three Sympatric Lizards in Response to Low-Temperature Stress.Animals (Basel). 2024 Apr 11;14(8):1158. doi: 10.3390/ani14081158. Animals (Basel). 2024. PMID: 38672309 Free PMC article.
-
Vertebrate sex determination: evolutionary plasticity of a fundamental switch.Nat Rev Genet. 2017 Nov;18(11):675-689. doi: 10.1038/nrg.2017.60. Epub 2017 Aug 14. Nat Rev Genet. 2017. PMID: 28804140 Review.
-
Post-Transcriptional Mechanisms Respond Rapidly to Ecologically Relevant Thermal Fluctuations During Temperature-Dependent Sex Determination.Integr Org Biol. 2020 Oct 7;2(1):obaa033. doi: 10.1093/iob/obaa033. eCollection 2020. Integr Org Biol. 2020. PMID: 33791571 Free PMC article.
-
Two transcriptionally distinct pathways drive female development in a reptile with both genetic and temperature dependent sex determination.PLoS Genet. 2021 Apr 15;17(4):e1009465. doi: 10.1371/journal.pgen.1009465. eCollection 2021 Apr. PLoS Genet. 2021. PMID: 33857129 Free PMC article.
References
-
- Bull JJ. Sex determination in reptiles. Q Rev Biol. 1980;55:3–21. doi: 10.1086/411613. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

