Chlorhexidine Induces VanA-Type Vancomycin Resistance Genes in Enterococci
- PMID: 26810654
- PMCID: PMC4808233
- DOI: 10.1128/AAC.02595-15
Chlorhexidine Induces VanA-Type Vancomycin Resistance Genes in Enterococci
Abstract
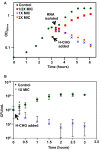
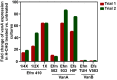
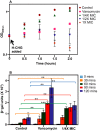
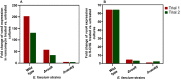
Chlorhexidine is a bisbiguanide antiseptic used for infection control. Vancomycin-resistantE. faecium(VREfm) is among the leading causes of hospital-acquired infections. VREfm may be exposed to chlorhexidine at supra- and subinhibitory concentrations as a result of chlorhexidine bathing and chlorhexidine-impregnated central venous catheter use. We used RNA sequencing to investigate how VREfm responds to chlorhexidine gluconate exposure. Among the 35 genes upregulated ≥10-fold after 15 min of exposure to the MIC of chlorhexidine gluconate were those encoding VanA-type vancomycin resistance (vanHAX) and those associated with reduced daptomycin susceptibility (liaXYZ). We confirmed thatvanAupregulation was not strain or species specific by querying other VanA-type VRE. VanB-type genes were not induced. ThevanHpromoter was found to be responsive to subinhibitory chlorhexidine gluconate in VREfm, as was production of the VanX protein. UsingvanHreporter experiments withBacillus subtilisand deletion analysis in VREfm, we found that this phenomenon is VanR dependent. Deletion ofvanRdid not result in increased chlorhexidine susceptibility, demonstrating thatvanHAXinduction is not protective against chlorhexidine. As expected, VanA-type VRE is more susceptible to ceftriaxone in the presence of sub-MIC chlorhexidine. Unexpectedly, VREfm is also more susceptible to vancomycin in the presence of subinhibitory chlorhexidine, suggesting that chlorhexidine-induced gene expression changes lead to additional alterations in cell wall synthesis. We conclude that chlorhexidine induces expression of VanA-type vancomycin resistance genes and genes associated with daptomycin nonsusceptibility. Overall, our results indicate that the impacts of subinhibitory chlorhexidine exposure on hospital-associated pathogens should be further investigated in laboratory studies.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Effect of Vancomycin on Cytoplasmic Peptidoglycan Intermediates and van Operon mRNA Levels in VanA-Type Vancomycin-Resistant Enterococcus faecium.J Bacteriol. 2021 Jul 22;203(16):e0023021. doi: 10.1128/JB.00230-21. Epub 2021 Jul 22. J Bacteriol. 2021. PMID: 34060906 Free PMC article.
-
Utilizing genomic analyses to investigate the first outbreak of vanA vancomycin-resistant Enterococcus in Australia with emergence of daptomycin non-susceptibility.J Med Microbiol. 2019 Mar;68(3):303-308. doi: 10.1099/jmm.0.000916. Epub 2019 Jan 21. J Med Microbiol. 2019. PMID: 30663951
-
Reduced Chlorhexidine and Daptomycin Susceptibility in Vancomycin-Resistant Enterococcus faecium after Serial Chlorhexidine Exposure.Antimicrob Agents Chemother. 2017 Dec 21;62(1):e01235-17. doi: 10.1128/AAC.01235-17. Print 2018 Jan. Antimicrob Agents Chemother. 2017. PMID: 29038276 Free PMC article.
-
Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: Towards a common nomenclature.Drug Resist Updat. 2018 Sep;40:25-39. doi: 10.1016/j.drup.2018.10.002. Epub 2018 Nov 2. Drug Resist Updat. 2018. PMID: 30447411 Review.
-
Acquired vancomycin resistance in clinically relevant pathogens.Future Microbiol. 2008 Oct;3(5):547-62. doi: 10.2217/17460913.3.5.547. Future Microbiol. 2008. PMID: 18811239 Review.
Cited by
-
Adaptation of the gut pathobiont Enterococcus faecalis to deoxycholate and taurocholate bile acids.Sci Rep. 2022 May 19;12(1):8485. doi: 10.1038/s41598-022-12552-3. Sci Rep. 2022. PMID: 35590028 Free PMC article.
-
Efficacy of Ceragenins in Controlling the Growth of Oral Microorganisms: Implications for Oral Hygiene Management.Pharmaceuticals (Basel). 2024 Feb 5;17(2):204. doi: 10.3390/ph17020204. Pharmaceuticals (Basel). 2024. PMID: 38399419 Free PMC article.
-
2CS-CHXT Operon Signature of Chlorhexidine Tolerance among Enterococcus faecium Isolates.Appl Environ Microbiol. 2019 Nov 14;85(23):e01589-19. doi: 10.1128/AEM.01589-19. Print 2019 Dec 1. Appl Environ Microbiol. 2019. PMID: 31562170 Free PMC article.
-
Subinhibitory Concentrations of Ciprofloxacin Enhance Antimicrobial Resistance and Pathogenicity of Enterococcus faecium.Antimicrob Agents Chemother. 2017 Apr 24;61(5):e02763-16. doi: 10.1128/AAC.02763-16. Print 2017 May. Antimicrob Agents Chemother. 2017. PMID: 28193670 Free PMC article.
-
Frequency of antiseptic resistance genes in clinical staphycocci and enterococci isolates in Turkey.Antimicrob Resist Infect Control. 2017 Aug 30;6:88. doi: 10.1186/s13756-017-0244-6. eCollection 2017. Antimicrob Resist Infect Control. 2017. PMID: 28861267 Free PMC article.
References
-
- Agudelo Higuita NI, Huycke MM. 2014. Enterococcal disease, epidemiology, and implications for treatment, p 45–70. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA.
-
- Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S, National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities. 2013. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect Control Hosp Epidemiol 34:1–14. doi:10.1086/668770. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

