PqsBC, a Condensing Enzyme in the Biosynthesis of the Pseudomonas aeruginosa Quinolone Signal: CRYSTAL STRUCTURE, INHIBITION, AND REACTION MECHANISM
- PMID: 26811339
- PMCID: PMC4807248
- DOI: 10.1074/jbc.M115.708453
PqsBC, a Condensing Enzyme in the Biosynthesis of the Pseudomonas aeruginosa Quinolone Signal: CRYSTAL STRUCTURE, INHIBITION, AND REACTION MECHANISM
Abstract
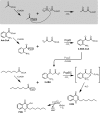
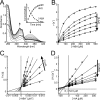
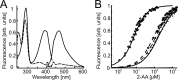
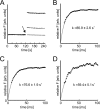
Pseudomonas aeruginosaproduces a number of alkylquinolone-type secondary metabolites best known for their antimicrobial effects and involvement in cell-cell communication. In the alkylquinolone biosynthetic pathway, the β-ketoacyl-(acyl carrier protein) synthase III (FabH)-like enzyme PqsBC catalyzes the condensation of octanoyl-coenzyme A and 2-aminobenzoylacetate (2-ABA) to form the signal molecule 2-heptyl-4(1H)-quinolone. PqsBC, a potential drug target, is unique for its heterodimeric arrangement and an active site different from that of canonical FabH-like enzymes. Considering the sequence dissimilarity between the subunits, a key question was how the two subunits are organized with respect to the active site. In this study, the PqsBC structure was determined to a 2 Å resolution, revealing that PqsB and PqsC have a pseudo-2-fold symmetry that unexpectedly mimics the FabH homodimer. PqsC has an active site composed of Cys-129 and His-269, and the surrounding active site cleft is hydrophobic in character and approximately twice the volume of related FabH enzymes that may be a requirement to accommodate the aromatic substrate 2-ABA. From physiological and kinetic studies, we identified 2-aminoacetophenone as a pathway-inherent competitive inhibitor of PqsBC, whose fluorescence properties could be used forin vitrobinding studies. In a time-resolved setup, we demonstrated that the catalytic histidine is not involved in acyl-enzyme formation, but contributes to an acylation-dependent increase in affinity for the second substrate 2-ABA. Introduction of Asn into the PqsC active site led to significant activity toward the desamino substrate analog benzoylacetate, suggesting that the substrate 2-ABA itself supplies the asparagine-equivalent amino function that assists in catalysis.
Keywords: 2-alkyl-4(1H)-quinolones; FabH; Pseudomonas aeruginosa (P. aeruginosa); Pseudomonas quinolone signal; biosynthesis; condensing enzyme; crystal structure; quorum sensing; secondary metabolism.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- Lyczak J. B., Cannon C. L., and Pier G. B. (2000) Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2, 1051–1060 - PubMed
-
- Williams P. (2007) Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153, 3923–3938 - PubMed
-
- Storz M. P., Maurer C. K., Zimmer C., Wagner N., Brengel C., de Jong J. C., Lucas S., Müsken M., Häussler S., Steinbach A., and Hartmann R. W. (2012) Validation of PqsD as an anti-biofilm target in Pseudomonas aeruginosa by development of small-molecule inhibitors. J. Am. Chem. Soc. 134, 16143–16146 - PubMed
-
- Lu C., Maurer C. K., Kirsch B., Steinbach A., and Hartmann R. W. (2014) Overcoming the unexpected functional inversion of a PqsR antagonist in Pseudomonas aeruginosa: an in vivo potent antivirulence agent targeting pqs quorum sensing. Angew. Chem. Int. Ed. Engl. 53, 1109–1112 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

