Life history effects on the molecular clock of autosomes and sex chromosomes
- PMID: 26811451
- PMCID: PMC4760823
- DOI: 10.1073/pnas.1515798113
Life history effects on the molecular clock of autosomes and sex chromosomes
Abstract
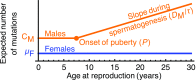
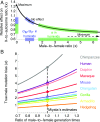
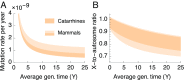
One of the foundational results in molecular evolution is that the rate at which neutral substitutions accumulate on a lineage equals the rate at which mutations arise. Traits that affect rates of mutation therefore also affect the phylogenetic "molecular clock." We consider the effects of sex-specific generation times and mutation rates in species with two sexes. In particular, we focus on the effects that the age of onset of male puberty and rates of spermatogenesis have likely had in hominids (great apes), considering a model that approximates features of the mutational process in mammals, birds, and some other vertebrates. As we show, this model can account for a number of seemingly disparate observations: notably, the puzzlingly low X-to-autosome ratios of substitution rates in humans and chimpanzees and differences in rates of autosomal substitutions among hominine lineages (i.e., humans, chimpanzees, and gorillas). The model further suggests how to translate pedigree-based estimates of human mutation rates into split times among extant hominoids (apes), given sex-specific life histories. In so doing, it largely bridges the gap reported between estimates of split times based on fossil and molecular evidence, in particular suggesting that the human-chimpanzee split may have occurred as recently as 6.6 Mya. The model also implies that the "generation time effect" should be stronger in short-lived species, explaining why the generation time has a major influence on yearly substitution rates in mammals but only a subtle one in human pedigrees.
Keywords: generation time effect; human–chimpanzee split; male mutation bias; molecular clock; mutational slowdown.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
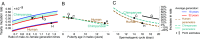
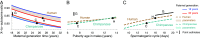

References
-
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge Univ Press; London: 1983.
-
- Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat. 2003;21(1):12–27. - PubMed
-
- Ségurel L, Wyman MJ, Przeworski M. Determinants of mutation rate variation in the human germline. Annu Rev Genomics Hum Genet. 2014;15:47–70. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

