Successful immunotherapy induces previously unidentified allergen-specific CD4+ T-cell subsets
- PMID: 26811452
- PMCID: PMC4780622
- DOI: 10.1073/pnas.1520180113
Successful immunotherapy induces previously unidentified allergen-specific CD4+ T-cell subsets
Abstract
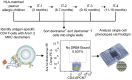
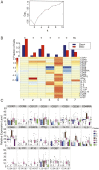
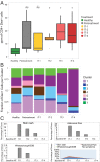
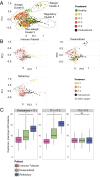
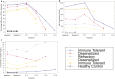
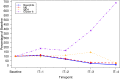
Allergen immunotherapy can desensitize even subjects with potentially lethal allergies, but the changes induced in T cells that underpin successful immunotherapy remain poorly understood. In a cohort of peanut-allergic participants, we used allergen-specific T-cell sorting and single-cell gene expression to trace the transcriptional "roadmap" of individual CD4+ T cells throughout immunotherapy. We found that successful immunotherapy induces allergen-specific CD4+ T cells to expand and shift toward an "anergic" Th2 T-cell phenotype largely absent in both pretreatment participants and healthy controls. These findings show that sustained success, even after immunotherapy is withdrawn, is associated with the induction, expansion, and maintenance of immunotherapy-specific memory and naive T-cell phenotypes as early as 3 mo into immunotherapy. These results suggest an approach for immune monitoring participants undergoing immunotherapy to predict the success of future treatment and could have implications for immunotherapy targets in other diseases like cancer, autoimmune disease, and transplantation.
Keywords: T cells; anergy; gene expression; immunotherapy; tolerance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Characterization of CD4+ T cell subsets in allergy.Curr Opin Immunol. 2012 Dec;24(6):700-6. doi: 10.1016/j.coi.2012.07.009. Epub 2012 Aug 10. Curr Opin Immunol. 2012. PMID: 22889592 Free PMC article. Review.
-
High dose allergen stimulation of T cells from house dust mite-allergic subjects induces expansion of IFN-gamma+ T Cells, apoptosis of CD4+IL-4+ T cells and T cell anergy.Int Arch Allergy Immunol. 2004 Jan;133(1):1-13. doi: 10.1159/000075248. Epub 2003 Nov 28. Int Arch Allergy Immunol. 2004. PMID: 14646373
-
Triggering of specific Toll-like receptors and proinflammatory cytokines breaks allergen-specific T-cell tolerance in human tonsils and peripheral blood.J Allergy Clin Immunol. 2013 Mar;131(3):875-85. doi: 10.1016/j.jaci.2012.10.051. Epub 2012 Dec 23. J Allergy Clin Immunol. 2013. PMID: 23265862
-
Allergen-specific CD4+ T cell responses in peripheral blood do not predict the early onset of clinical efficacy during grass pollen sublingual immunotherapy.Clin Exp Allergy. 2012 Dec;42(12):1745-55. doi: 10.1111/cea.12015. Clin Exp Allergy. 2012. PMID: 23181790 Clinical Trial.
-
Specific immunotherapy and turning off the T cell: how does it work?Ann Allergy Asthma Immunol. 2011 Nov;107(5):381-92. doi: 10.1016/j.anai.2011.05.017. Epub 2011 Jun 25. Ann Allergy Asthma Immunol. 2011. PMID: 22018608 Review.
Cited by
-
Peanut oral immunotherapy differentially suppresses clonally distinct subsets of T helper cells.J Clin Invest. 2022 Jan 18;132(2):e150634. doi: 10.1172/JCI150634. J Clin Invest. 2022. PMID: 34813505 Free PMC article. Clinical Trial.
-
Deciphering the black box of food allergy mechanisms.Ann Allergy Asthma Immunol. 2017 Jan;118(1):21-27. doi: 10.1016/j.anai.2016.10.017. Ann Allergy Asthma Immunol. 2017. PMID: 28007085 Free PMC article. Review.
-
Biomarkers in oral immunotherapy.J Zhejiang Univ Sci B. 2022 Sept 15;23(9):705-731. doi: 10.1631/jzus.B2200047. J Zhejiang Univ Sci B. 2022. PMID: 36111569 Free PMC article. Review.
-
Emerging Food Allergy Biomarkers.J Allergy Clin Immunol Pract. 2020 Sep;8(8):2516-2524. doi: 10.1016/j.jaip.2020.04.054. J Allergy Clin Immunol Pract. 2020. PMID: 32888527 Free PMC article. Review.
-
Treating cat allergy with monoclonal IgG antibodies that bind allergen and prevent IgE engagement.Nat Commun. 2018 Apr 12;9(1):1421. doi: 10.1038/s41467-018-03636-8. Nat Commun. 2018. PMID: 29650949 Free PMC article. Clinical Trial.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

