Proinflammatory signal suppresses proliferation and shifts macrophage metabolism from Myc-dependent to HIF1α-dependent
- PMID: 26811453
- PMCID: PMC4760828
- DOI: 10.1073/pnas.1518000113
Proinflammatory signal suppresses proliferation and shifts macrophage metabolism from Myc-dependent to HIF1α-dependent
Abstract
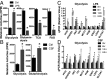
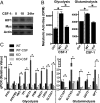
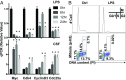
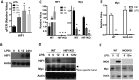
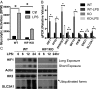
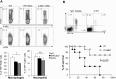
As a phenotypically plastic cellular population, macrophages change their physiology in response to environmental signals. Emerging evidence suggests that macrophages are capable of tightly coordinating their metabolic programs to adjust their immunological and bioenergetic functional properties, as needed. Upon mitogenic stimulation, quiescent macrophages enter the cell cycle, increasing their bioenergetic and biosynthetic activity to meet the demands of cell growth. Proinflammatory stimulation, however, suppresses cell proliferation, while maintaining a heightened metabolic activity imposed by the production of bactericidal factors. Here, we report that the mitogenic stimulus, colony-stimulating factor 1 (CSF-1), engages a myelocytomatosis viral oncogen (Myc)-dependent transcriptional program that is responsible for cell cycle entry and the up-regulation of glucose and glutamine catabolism in bone marrow-derived macrophages (BMDMs). However, the proinflammatory stimulus, lipopolysaccharide (LPS), suppresses Myc expression and cell proliferation and engages a hypoxia-inducible factor alpha (HIF1α)-dependent transcriptional program that is responsible for heightened glycolysis. The acute deletion of Myc or HIF1α selectively impaired the CSF-1- or LPS-driven metabolic activities in BMDM, respectively. Finally, inhibition of glycolysis by 2-deoxyglucose (2-DG) or genetic deletion of HIF1α suppressed LPS-induced inflammation in vivo. Our studies indicate that a switch from a Myc-dependent to a HIF1α-dependent transcriptional program may regulate the robust bioenergetic support for an inflammatory response, while sparing Myc-dependent proliferation.
Keywords: HIF1α; Myc; cell cycle; macrophage; metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Ginhoux F, Jung S. Monocytes and macrophages: Developmental pathways and tissue homeostasis. Nat Rev Immunol. 2014;14(6):392–404. - PubMed
-
- Sieweke MH, Allen JE. Beyond stem cells: Self-renewal of differentiated macrophages. Science. 2013;342(6161):1242974. - PubMed
-
- Newsholme EA, Crabtree B, Ardawi MS. The role of high rates of glycolysis and glutamine utilization in rapidly dividing cells. Biosci Rep. 1985;5(5):393–400. - PubMed
-
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. - PubMed
-
- Shaw RJ. Glucose metabolism and cancer. Curr Opin Cell Biol. 2006;18(6):598–608. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

