Molecular Basis of Orb2 Amyloidogenesis and Blockade of Memory Consolidation
- PMID: 26812143
- PMCID: PMC4727891
- DOI: 10.1371/journal.pbio.1002361
Molecular Basis of Orb2 Amyloidogenesis and Blockade of Memory Consolidation
Abstract
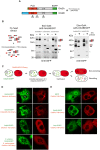
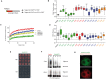
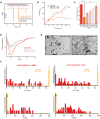
Amyloids are ordered protein aggregates that are typically associated with neurodegenerative diseases and cognitive impairment. By contrast, the amyloid-like state of the neuronal RNA binding protein Orb2 in Drosophila was recently implicated in memory consolidation, but it remains unclear what features of this functional amyloid-like protein give rise to such diametrically opposed behaviour. Here, using an array of biophysical, cell biological and behavioural assays we have characterized the structural features of Orb2 from the monomer to the amyloid state. Surprisingly, we find that Orb2 shares many structural traits with pathological amyloids, including the intermediate toxic oligomeric species, which can be sequestered in vivo in hetero-oligomers by pathological amyloids. However, unlike pathological amyloids, Orb2 rapidly forms amyloids and its toxic intermediates are extremely transient, indicating that kinetic parameters differentiate this functional amyloid from pathological amyloids. We also observed that a well-known anti-amyloidogenic peptide interferes with long-term memory in Drosophila. These results provide structural insights into how the amyloid-like state of the Orb2 protein can stabilize memory and be nontoxic. They also provide insight into how amyloid-based diseases may affect memory processes.
Conflict of interest statement
The authors declare competing financial interest: MCV and RH are co-inventors on an international patent application (reference EP15382176.4) covering the results contained in this article. Any potential income generated by exploitation of the patent rights and received by their employer, the CSIC, shall be shared with these authors according to Spanish law and the regulations of the CSIC.
Figures




References
-
- Chernoff YO. Amyloidogenic domains, prions and structural inheritance: rudiments of early life or recent acquisition? Curr Opin Chem Biol. 2004;8(6):665–71. - PubMed
-
- Lansbury PT, Lashuel HA. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443(7113):774–9. - PubMed
-
- Pepys MB. Amyloidosis. Annual review of medicine. 2006;57:223–41. - PubMed
-
- Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid—from bacteria to humans. Trends Biochem Sci. 2007;32(5):217–24. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

