Probing the Limits to MicroRNA-Mediated Control of Gene Expression
- PMID: 26812364
- PMCID: PMC4727922
- DOI: 10.1371/journal.pcbi.1004715
Probing the Limits to MicroRNA-Mediated Control of Gene Expression
Abstract
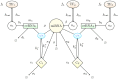
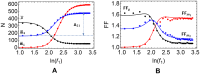
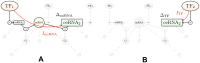
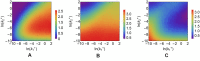
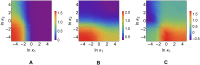
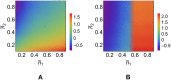
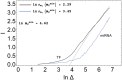
According to the 'ceRNA hypothesis', microRNAs (miRNAs) may act as mediators of an effective positive interaction between long coding or non-coding RNA molecules, carrying significant potential implications for a variety of biological processes. Here, inspired by recent work providing a quantitative description of small regulatory elements as information-conveying channels, we characterize the effectiveness of miRNA-mediated regulation in terms of the optimal information flow achievable between modulator (transcription factors) and target nodes (long RNAs). Our findings show that, while a sufficiently large degree of target derepression is needed to activate miRNA-mediated transmission, (a) in case of differential mechanisms of complex processing and/or transcriptional capabilities, regulation by a post-transcriptional miRNA-channel can outperform that achieved through direct transcriptional control; moreover, (b) in the presence of large populations of weakly interacting miRNA molecules the extra noise coming from titration disappears, allowing the miRNA-channel to process information as effectively as the direct channel. These observations establish the limits of miRNA-mediated post-transcriptional cross-talk and suggest that, besides providing a degree of noise buffering, this type of control may be effectively employed in cells both as a failsafe mechanism and as a preferential fine tuner of gene expression, pointing to the specific situations in which each of these functionalities is maximized.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Integrated analyses to reconstruct microRNA-mediated regulatory networks in mouse liver using high-throughput profiling.BMC Genomics. 2015;16 Suppl 2(Suppl 2):S12. doi: 10.1186/1471-2164-16-S2-S12. Epub 2015 Jan 21. BMC Genomics. 2015. PMID: 25707768 Free PMC article.
-
Kinetic Modelling of Competition and Depletion of Shared miRNAs by Competing Endogenous RNAs.Methods Mol Biol. 2019;1912:367-409. doi: 10.1007/978-1-4939-8982-9_15. Methods Mol Biol. 2019. PMID: 30635902 Review.
-
MicroRNA-mediated regulation in the mammalian circadian rhythm.J Theor Biol. 2012 Jul 7;304:103-10. doi: 10.1016/j.jtbi.2012.03.037. Epub 2012 Apr 8. J Theor Biol. 2012. PMID: 22554948
-
'Traffic light rules': Chromatin states direct miRNA-mediated network motifs running by integrating epigenome and regulatome.Biochim Biophys Acta. 2016 Jul;1860(7):1475-88. doi: 10.1016/j.bbagen.2016.04.008. Epub 2016 Apr 14. Biochim Biophys Acta. 2016. PMID: 27091612
-
The evolution of gene regulation by transcription factors and microRNAs.Nat Rev Genet. 2007 Feb;8(2):93-103. doi: 10.1038/nrg1990. Nat Rev Genet. 2007. PMID: 17230196 Review.
Cited by
-
Differential correlation analysis of glioblastoma reveals immune ceRNA interactions predictive of patient survival.BMC Bioinformatics. 2017 Feb 28;18(1):132. doi: 10.1186/s12859-017-1557-4. BMC Bioinformatics. 2017. PMID: 28241741 Free PMC article.
-
Competing endogenous RNA crosstalk at system level.PLoS Comput Biol. 2019 Nov 1;15(11):e1007474. doi: 10.1371/journal.pcbi.1007474. eCollection 2019 Nov. PLoS Comput Biol. 2019. PMID: 31675359 Free PMC article.
-
Out-of-Equilibrium ceRNA Crosstalk.Methods Mol Biol. 2025;2883:167-193. doi: 10.1007/978-1-0716-4290-0_8. Methods Mol Biol. 2025. PMID: 39702709
-
Molecular Network-Based Identification of Competing Endogenous RNAs in Thyroid Carcinoma.Genes (Basel). 2018 Jan 19;9(1):44. doi: 10.3390/genes9010044. Genes (Basel). 2018. PMID: 29351231 Free PMC article.
-
ceRNA crosstalk stabilizes protein expression and affects the correlation pattern of interacting proteins.Sci Rep. 2017 Mar 7;7:43673. doi: 10.1038/srep43673. Sci Rep. 2017. PMID: 28266541 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

