The Phenazine 2-Hydroxy-Phenazine-1-Carboxylic Acid Promotes Extracellular DNA Release and Has Broad Transcriptomic Consequences in Pseudomonas chlororaphis 30-84
- PMID: 26812402
- PMCID: PMC4727817
- DOI: 10.1371/journal.pone.0148003
The Phenazine 2-Hydroxy-Phenazine-1-Carboxylic Acid Promotes Extracellular DNA Release and Has Broad Transcriptomic Consequences in Pseudomonas chlororaphis 30-84
Abstract
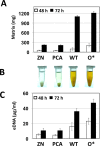
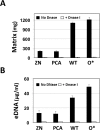
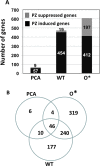
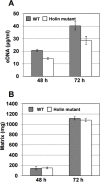
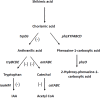
Enhanced production of 2-hydroxy-phenazine-1-carboxylic acid (2-OH-PCA) by the biological control strain Pseudomonas chlororaphis 30-84 derivative 30-84O* was shown previously to promote cell adhesion and alter the three-dimensional structure of surface-attached biofilms compared to the wild type. The current study demonstrates that production of 2-OH-PCA promotes the release of extracellular DNA, which is correlated with the production of structured biofilm matrix. Moreover, the essential role of the extracellular DNA in maintaining the mass and structure of the 30-84 biofilm matrix is demonstrated. To better understand the role of different phenazines in biofilm matrix production and gene expression, transcriptomic analyses were conducted comparing gene expression patterns of populations of wild type, 30-84O* and a derivative of 30-84 producing only PCA (30-84PCA) to a phenazine defective mutant (30-84ZN) when grown in static cultures. RNA-Seq analyses identified a group of 802 genes that were differentially expressed by the phenazine producing derivatives compared to 30-84ZN, including 240 genes shared by the two 2-OH-PCA producing derivatives, the wild type and 30-84O*. A gene cluster encoding a bacteriophage-derived pyocin and its lysis cassette was upregulated in 2-OH-PCA producing derivatives. A holin encoded in this gene cluster was found to contribute to the release of eDNA in 30-84 biofilm matrices, demonstrating that the influence of 2-OH-PCA on eDNA production is due in part to cell autolysis as a result of pyocin production and release. The results expand the current understanding of the functions different phenazines play in the survival of bacteria in biofilm-forming communities.
Conflict of interest statement
Figures
References
-
- Haas D, Defago G (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3: 307–319. - PubMed
-
- Mavrodi DV, Blankenfeldt W, Thomashow LS (2006) Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol 44: 417–445. - PubMed
-
- Pierson EA, Wang D, Pierson LS III (2013) Roles and Regulation of Phenazines in the Biological Control Strain Pseudomonas chlororaphis 30–84 In: Chincholkar S and Thomashow L, eds. Microbial Phenazines. Springer; Berlin Heidelberg: pp. 141–162.
-
- Pierson LS III, Thomashow LS (1992) Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30–84. Mol Plant-Microbe Interact 5: 330–339. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

