Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome
- PMID: 26812545
- PMCID: PMC4764553
- DOI: 10.7554/eLife.11911
Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome
Erratum in
-
Correction: Human cytomegalovirus IE1 alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome.Elife. 2016 Mar 14;5:e15893. doi: 10.7554/eLife.15893. Elife. 2016. PMID: 26974472 Free PMC article. No abstract available.
Abstract
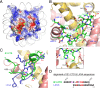
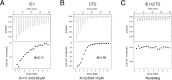
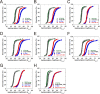
Human cytomegalovirus (hCMV) immediate early 1 (IE1) protein associates with condensed chromatin of the host cell during mitosis. We have determined the structure of the chromatin-tethering domain (CTD) of IE1 bound to the nucleosome core particle, and discovered that IE1-CTD specifically interacts with the H2A-H2B acidic patch and impairs the compaction of higher-order chromatin structure. Our results suggest that IE1 loosens up the folding of host chromatin during hCMV infections.
Keywords: biochemistry; biophysics; chromatin; crystal structure; cytomegalovirus; human; nucleosome; structural biology; virus.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
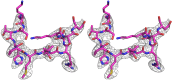


References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX : a comprehensive python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Ahn J-H, Brignole EJ, Hayward GS. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Molecular and Cellular Biology. 1998;18:4899–4913. doi: 10.1128/MCB.18.8.4899. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

