Genetic Variation, Not Cell Type of Origin, Underlies the Majority of Identifiable Regulatory Differences in iPSCs
- PMID: 26812582
- PMCID: PMC4727884
- DOI: 10.1371/journal.pgen.1005793
Genetic Variation, Not Cell Type of Origin, Underlies the Majority of Identifiable Regulatory Differences in iPSCs
Abstract
The advent of induced pluripotent stem cells (iPSCs) revolutionized human genetics by allowing us to generate pluripotent cells from easily accessible somatic tissues. This technology can have immense implications for regenerative medicine, but iPSCs also represent a paradigm shift in the study of complex human phenotypes, including gene regulation and disease. Yet, an unresolved caveat of the iPSC model system is the extent to which reprogrammed iPSCs retain residual phenotypes from their precursor somatic cells. To directly address this issue, we used an effective study design to compare regulatory phenotypes between iPSCs derived from two types of commonly used somatic precursor cells. We find a remarkably small number of differences in DNA methylation and gene expression levels between iPSCs derived from different somatic precursors. Instead, we demonstrate genetic variation is associated with the majority of identifiable variation in DNA methylation and gene expression levels. We show that the cell type of origin only minimally affects gene expression levels and DNA methylation in iPSCs, and that genetic variation is the main driver of regulatory differences between iPSCs of different donors. Our findings suggest that studies using iPSCs should focus on additional individuals rather than clones from the same individual.
Conflict of interest statement
JKP is on the advisory board of 23andMe with stock options. No other competing interests exist.
Figures
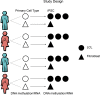

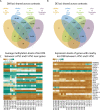

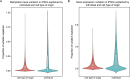
Comment in
-
STEM CELLS. Reprogrammed cells leave their past lives behind.Nat Methods. 2016 Apr;13(4):292. doi: 10.1038/nmeth.3829. Nat Methods. 2016. PMID: 27482572 No abstract available.
References
-
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, et al. (2007) Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 131: 861–872. - PubMed
-
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, et al. (2007) Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science 318: 1917–1920. - PubMed
-
- Yamanaka S, Takahashi K (2006) Induction of pluripotent stem cells from mouse fibroblast cultures. Tanpakushitsu Kakusan Koso 51: 2346–2351. - PubMed
-
- Okita K, Ichisaka T, Yamanaka S (2007) Generation of germline-competent induced pluripotent stem cells. Nature 448: 313–317. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

