Splicing factor mutations predict poor prognosis in patients with de novo acute myeloid leukemia
- PMID: 26812887
- PMCID: PMC4891028
- DOI: 10.18632/oncotarget.7000
Splicing factor mutations predict poor prognosis in patients with de novo acute myeloid leukemia
Abstract
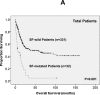
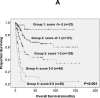
Mutations in splicing factor (SF) genes are frequently detected in myelodysplastic syndrome, but the prognostic relevance of these genes mutations in acute myeloid leukemia (AML) remains unclear. In this study, we investigated mutations of three SF genes, SF3B1, U2AF1 and SRSF2, by Sanger sequencing in 500 patients with de novo AML and analysed their clinical relevance. SF mutations were identified in 10.8% of total cohort and 13.2% of those with intermediate-risk cytogenetics. SF mutations were closely associated with RUNX1, ASXL1, IDH2 and TET2 mutations. SF-mutated AML patients had a significantly lower complete remission rate and shorter disease-free survival (DFS) and overall survival (OS) than those without the mutation. Multivariate analysis demonstrated that SFmutation was an independent poor prognostic factor for DFS and OS. A scoring system incorporating SF mutation and ten other prognostic factors was proved very useful to risk-stratify AML patients. Sequential study of paired samples showed that SF mutations were stable during AML evolution. In conclusion, SF mutations are associated with distinct clinic-biological features and poor prognosis in de novo AML patients and are rather stable during disease progression. These mutations may be potential targets for novel treatment and biomarkers for disease monitoring in AML.
Keywords: de novo AML; paired sample; prognosis; splicing factor mutations.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






References
-
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. - PubMed
-
- Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, Godfrey AL, Rapado I, Cvejic A, Rance R, McGee C, Ellis P, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. - PMC - PubMed
-
- Thol F, Kade S, Schlarmann C, Loffeld P, Morgan M, Krauter J, Wlodarski MW, Kolking B, Wichmann M, Gorlich K, Gohring G, Bug G, Ottmann O, Niemeyer CM, Hofmann WK, Schlegelberger B, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119:3578–3584. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

