A cotton miRNA is involved in regulation of plant response to salt stress
- PMID: 26813144
- PMCID: PMC4728436
- DOI: 10.1038/srep19736
A cotton miRNA is involved in regulation of plant response to salt stress
Abstract
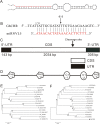
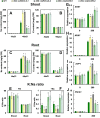
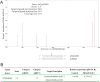
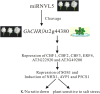
The present study functionally identified a new microRNA (microRNA ovual line 5, miRNVL5) with its target gene GhCHR from cotton (Gossypium hirsutum). The sequence of miRNVL5 precursor is 104 nt long, with a well developed secondary structure. GhCHR contains two DC1 and three PHD Cys/His-rich domains, suggesting that GhCHR encodes a zinc-finger domain-containing transcription factor. miRNVL5 and GhCHR express at various developmental stages of cotton. Under salt stress (50-400 mM NaCl), miRNVL5 expression was repressed, with concomitant high expression of GhCHR in cotton seedlings. Ectopic expression of GhCHR in Arabidopsis conferred salt stress tolerance by reducing Na(+) accumulation in plants and improving primary root growth and biomass. Interestingly, Arabidopsis constitutively expressing miRNVL5 showed hypersensitivity to salt stress. A GhCHR orthorlous gene At2g44380 from Arabidopsis that can be cleaved by miRNVL5 was identified by degradome sequencing, but no confidential miRNVL5 homologs in Arabidopsis have been identified. Microarray analysis of miRNVL5 transgenic Arabidopsis showed six downstream genes (CBF1, CBF2, CBF3, ERF4, AT3G22920, and AT3G49200), which were induced by salt stress in wild-type but repressed in miRNVL5-expressing Arabidopsis. These results indicate that miRNVL5 is involved in regulation of plant response to salt stress.
Figures




Similar articles
-
Overexpression of a cotton (Gossypium hirsutum) WRKY gene, GhWRKY34, in Arabidopsis enhances salt-tolerance of the transgenic plants.Plant Physiol Biochem. 2015 Nov;96:311-20. doi: 10.1016/j.plaphy.2015.08.016. Epub 2015 Aug 28. Plant Physiol Biochem. 2015. PMID: 26332661
-
Comprehensive analysis of differentially expressed genes and transcriptional regulation induced by salt stress in two contrasting cotton genotypes.BMC Genomics. 2014 Sep 5;15(1):760. doi: 10.1186/1471-2164-15-760. BMC Genomics. 2014. PMID: 25189468 Free PMC article.
-
GhSOS1, a plasma membrane Na+/H+ antiporter gene from upland cotton, enhances salt tolerance in transgenic Arabidopsis thaliana.PLoS One. 2017 Jul 19;12(7):e0181450. doi: 10.1371/journal.pone.0181450. eCollection 2017. PLoS One. 2017. PMID: 28723926 Free PMC article.
-
Roles of S-Adenosylmethionine and Its Derivatives in Salt Tolerance of Cotton.Int J Mol Sci. 2023 May 30;24(11):9517. doi: 10.3390/ijms24119517. Int J Mol Sci. 2023. PMID: 37298464 Free PMC article. Review.
-
Advances in the regulation of plant salt-stress tolerance by miRNA.Mol Biol Rep. 2022 Jun;49(6):5041-5055. doi: 10.1007/s11033-022-07179-6. Epub 2022 Apr 5. Mol Biol Rep. 2022. PMID: 35381964 Review.
Cited by
-
miRNAs Are Involved in Determining the Improved Vigor of Autotetrapoid Chrysanthemum nankingense.Front Plant Sci. 2016 Sep 28;7:1412. doi: 10.3389/fpls.2016.01412. eCollection 2016. Front Plant Sci. 2016. PMID: 27733854 Free PMC article.
-
microRNAs as reference genes for quantitative PCR in cotton.PLoS One. 2017 Apr 17;12(4):e0174722. doi: 10.1371/journal.pone.0174722. eCollection 2017. PLoS One. 2017. PMID: 28414734 Free PMC article.
-
Genome-wide Characterization of the JmjC Domain-Containing Histone Demethylase Gene Family Reveals GhJMJ24 and GhJMJ49 Involving in Somatic Embryogenesis Process in Cotton.Front Mol Biosci. 2022 Apr 27;9:888983. doi: 10.3389/fmolb.2022.888983. eCollection 2022. Front Mol Biosci. 2022. PMID: 35573733 Free PMC article.
-
Salt stress-induced FERROCHELATASE 1 improves resistance to salt stress by limiting sodium accumulation in Arabidopsis thaliana.Sci Rep. 2017 Nov 7;7(1):14737. doi: 10.1038/s41598-017-13593-9. Sci Rep. 2017. PMID: 29116128 Free PMC article.
-
Integration of mRNA and miRNA Analysis Reveals the Regulation of Salt Stress Response in Rapeseed (Brassica napus L.).Plants (Basel). 2025 Aug 4;14(15):2418. doi: 10.3390/plants14152418. Plants (Basel). 2025. PMID: 40805768 Free PMC article.
References
-
- Türkan I. & Demiral T. Recent developments in understanding salinity tolerance. Environ. Exp. Bot. 67, 2–9 (2009).
-
- Munns R. & Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681 (2008). - PubMed
-
- Zhu J. K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6, 441–445 (2003). - PubMed
-
- De Costa W., Zorb C., Hautung W. & Schubert S. Salt resistance is determined by osmotic adjustment and abscisic acid in newly developed maize hybrids in the first phase of salt stress. Physiol. Plant 131, 311–321 (2007). - PubMed
-
- Jones-Rhoades M. W. & Bartel D. P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell, 14, 787–799 (2004). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

