Functional analysis of androgen receptor mutations that confer anti-androgen resistance identified in circulating cell-free DNA from prostate cancer patients
- PMID: 26813233
- PMCID: PMC4729137
- DOI: 10.1186/s13059-015-0864-1
Functional analysis of androgen receptor mutations that confer anti-androgen resistance identified in circulating cell-free DNA from prostate cancer patients
Abstract
Background: The androgen receptor (AR) is a pivotal drug target for the treatment of prostate cancer, including its lethal castration-resistant (CRPC) form. All current non-steroidal AR antagonists, such as hydroxyflutamide, bicalutamide, and enzalutamide, target the androgen binding site of the receptor, competing with endogenous androgenic steroids. Several AR mutations in this binding site have been associated with poor prognosis and resistance to conventional prostate cancer drugs. In order to develop an effective CRPC therapy, it is crucial to understand the effects of these mutations on the functionality of the AR and its ability to interact with endogenous steroids and conventional AR inhibitors.
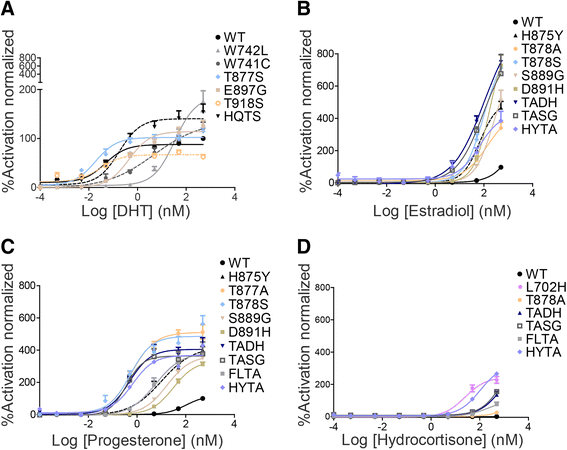
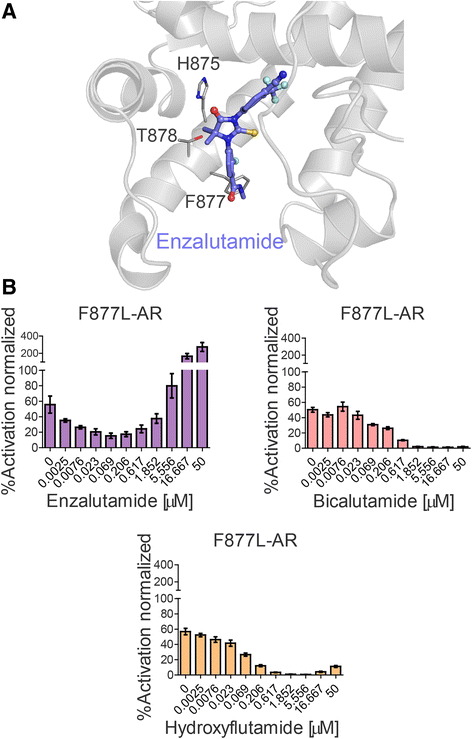
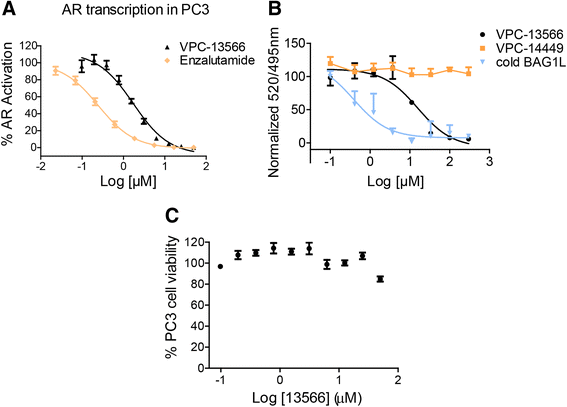
Results: We previously utilized circulating cell-free DNA (cfDNA) sequencing technology to examine the AR gene for the presence of mutations in CRPC patients. By modifying our sequencing and data analysis approaches, we identify four additional single AR mutations and five mutation combinations associated with CRPC. Importantly, we conduct experimental functionalization of all the AR mutations identified by the current and previous cfDNA sequencing to reveal novel gain-of-function scenarios. Finally, we evaluate the effect of a novel class of AR inhibitors targeting the binding function 3 (BF3) site on the activity of CRPC-associated AR mutants.
Conclusions: This work demonstrates the feasibility of a prognostic and/or diagnostic platform combining the direct identification of AR mutants from patients' serum, and the functional characterization of these mutants in order to provide personalized recommendations regarding the best future therapy.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

