One ring to rule them all: Current trends in combating bacterial resistance to the β-lactams
- PMID: 26813250
- PMCID: PMC4941212
- DOI: 10.1002/pro.2889
One ring to rule them all: Current trends in combating bacterial resistance to the β-lactams
Abstract
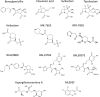
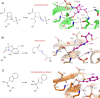
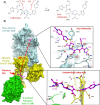
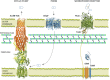
From humble beginnings of a contaminated petri dish, β-lactam antibiotics have distinguished themselves among some of the most powerful drugs in human history. The devastating effects of antibiotic resistance have nevertheless led to an "arms race" with disquieting prospects. The emergence of multidrug resistant bacteria threatens an ever-dwindling antibiotic arsenal, calling for new discovery, rediscovery, and innovation in β-lactam research. Here the current state of β-lactam antibiotics from a structural perspective was reviewed.
Keywords: multidrug resistant bacteria; penicillin-binding protein; resistance; β-lactam; β-lactamase.
© 2016 The Protein Society.
Figures

References
-
- Lewis K (2013) Platforms for antibiotic discovery. Nat Rev Drug Discov 12:371–387. - PubMed
-
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J (2009) Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. - PubMed
-
- Abraham EP, Chain E (1988) An enzyme from bacteria able to destroy penicillin. 1940. Rev Infect Dis 10:677–678. - PubMed
-
- Bush K, Fisher JF (2011) Epidemiological expansion, structural studies, and clinical challenges of new β‐lactamases from Gram‐negative bacteria. Annu Rev Microbiol 65:455–478. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

