A survey of best practices for RNA-seq data analysis
- PMID: 26813401
- PMCID: PMC4728800
- DOI: 10.1186/s13059-016-0881-8
A survey of best practices for RNA-seq data analysis
Erratum in
-
Erratum to: A survey of best practices for RNA-seq data analysis.Genome Biol. 2016 Aug 26;17(1):181. doi: 10.1186/s13059-016-1047-4. Genome Biol. 2016. PMID: 27565134 Free PMC article. No abstract available.
Abstract
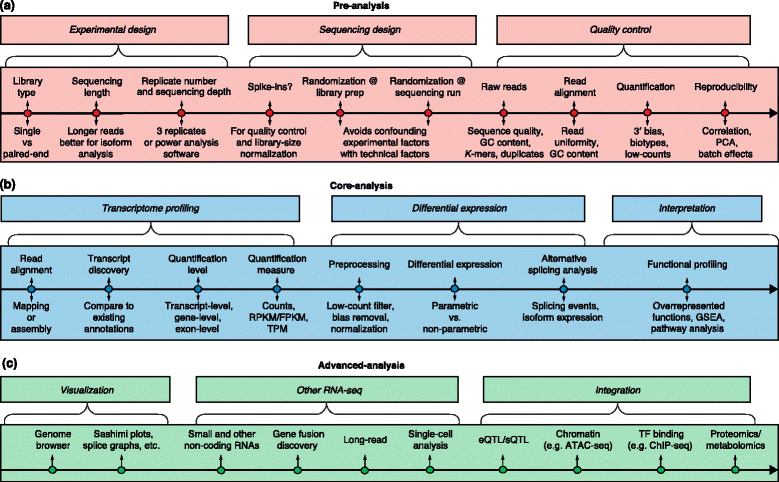
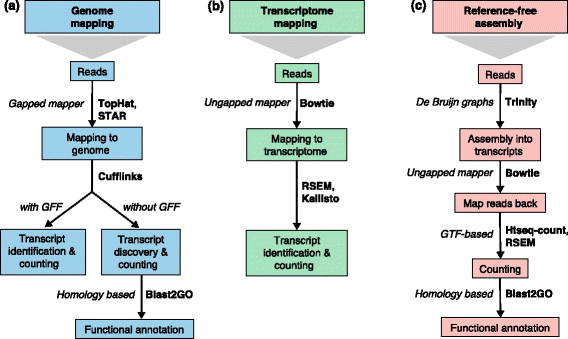
RNA-sequencing (RNA-seq) has a wide variety of applications, but no single analysis pipeline can be used in all cases. We review all of the major steps in RNA-seq data analysis, including experimental design, quality control, read alignment, quantification of gene and transcript levels, visualization, differential gene expression, alternative splicing, functional analysis, gene fusion detection and eQTL mapping. We highlight the challenges associated with each step. We discuss the analysis of small RNAs and the integration of RNA-seq with other functional genomics techniques. Finally, we discuss the outlook for novel technologies that are changing the state of the art in transcriptomics.
Figures