Combined Chlorophyll Fluorescence and Transcriptomic Analysis Identifies the P3/P4 Transition as a Key Stage in Rice Leaf Photosynthetic Development
- PMID: 26813793
- PMCID: PMC4775128
- DOI: 10.1104/pp.15.01624
Combined Chlorophyll Fluorescence and Transcriptomic Analysis Identifies the P3/P4 Transition as a Key Stage in Rice Leaf Photosynthetic Development
Abstract
Leaves are derived from heterotrophic meristem tissue that, at some point, must make the transition to autotrophy via the initiation of photosynthesis. However, the timing and spatial coordination of the molecular and cellular processes underpinning this switch are poorly characterized. Here, we report on the identification of a specific stage in rice (Oryza sativa) leaf development (P3/P4 transition) when photosynthetic competence is first established. Using a combined physiological and molecular approach, we show that elements of stomatal and vascular differentiation are coordinated with the onset of measurable light absorption for photosynthesis. Moreover, by exploring the response of the system to environmental perturbation, we show that the earliest stages of rice leaf development have significant plasticity with respect to elements of cellular differentiation of relevance for mature leaf photosynthetic performance. Finally, by performing an RNA sequencing analysis targeted at the early stages of rice leaf development, we uncover a palette of genes whose expression likely underpins the acquisition of photosynthetic capability. Our results identify the P3/P4 transition as a highly dynamic stage in rice leaf development when several processes for the initiation of photosynthetic competence are coordinated. As well as identifying gene targets for future manipulation of rice leaf structure/function, our data highlight a developmental window during which such manipulations are likely to be most effective.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures

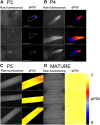
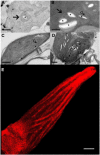

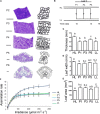
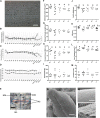

References
-
- Amiour N, Imbaud S, Clément G, Agier N, Zivy M, Valot B, Balliau T, Armengaud P, Quilleré I, Cañas R, et al. (2012) The use of metabolomics integrated with transcriptomic and proteomic studies for identifying key steps involved in the control of nitrogen metabolism in crops such as maize. J Exp Bot 63: 5017–5033 - PubMed
-
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GTS, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 - PubMed
-
- Arvidsson S, Kwasniewski M, Riano-Pachon DM, Mueller-Roeber B (2008) QuantPrime – a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics, 9: DOI: 10.1186/1471-2105-9-465 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

